Deposition Date
2018-01-08
Release Date
2018-02-21
Last Version Date
2024-10-30
Entry Detail
PDB ID:
6C2I
Keywords:
Title:
Structure of Bace-1 (Beta-Secretase) in complex with : N-(3-((1R,5S,6R)-3-amino-5-methyl-2-oxa-4-azabicyclo[4.1.0]hept-3-en-5-yl)-4-fluorophenyl)-5-methoxypyrazine-2-carboxamide
Biological Source:
Source Organism(s):
Homo sapiens (Taxon ID: 9606)
Expression System(s):
Method Details:
Experimental Method:
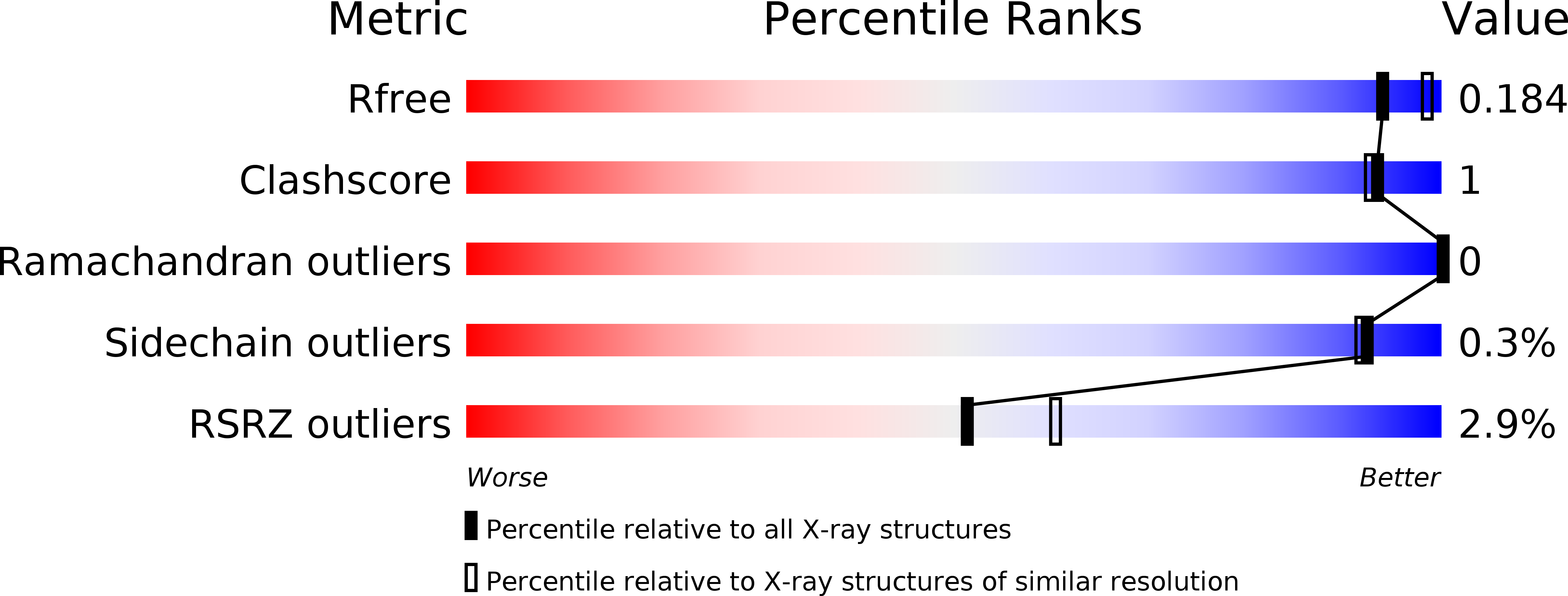
Resolution:
1.95 Å
R-Value Free:
0.18
R-Value Work:
0.15
R-Value Observed:
0.15
Space Group:
P 61 2 2