Deposition Date
2017-10-27
Release Date
2018-09-26
Last Version Date
2023-10-04
Entry Detail
PDB ID:
6BG5
Keywords:
Title:
Structure of 1-(benzo[d][1,3]dioxol-5-ylmethyl)-1-(1-propylpiperidin-4-yl)-3-(3-(trifluoromethyl)phenyl)urea bound to DCN1
Biological Source:
Source Organism:
Enterobacteria phage T4 (Taxon ID: 10665)
Homo sapiens (Taxon ID: 9606)
Homo sapiens (Taxon ID: 9606)
Host Organism:
Method Details:
Experimental Method:
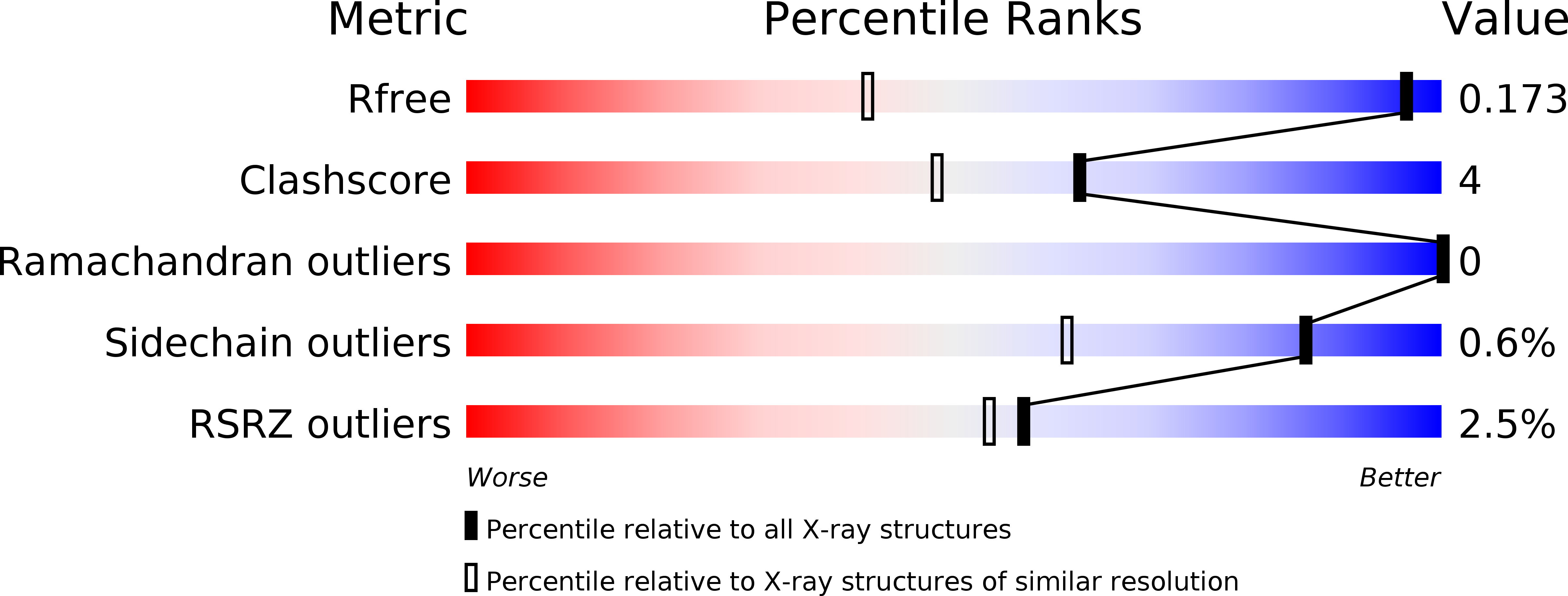
Resolution:
1.10 Å
R-Value Free:
0.17
R-Value Work:
0.14
R-Value Observed:
0.14
Space Group:
P 1 21 1