Deposition Date
2017-10-23
Release Date
2018-05-16
Last Version Date
2023-10-25
Entry Detail
PDB ID:
6BDO
Keywords:
Title:
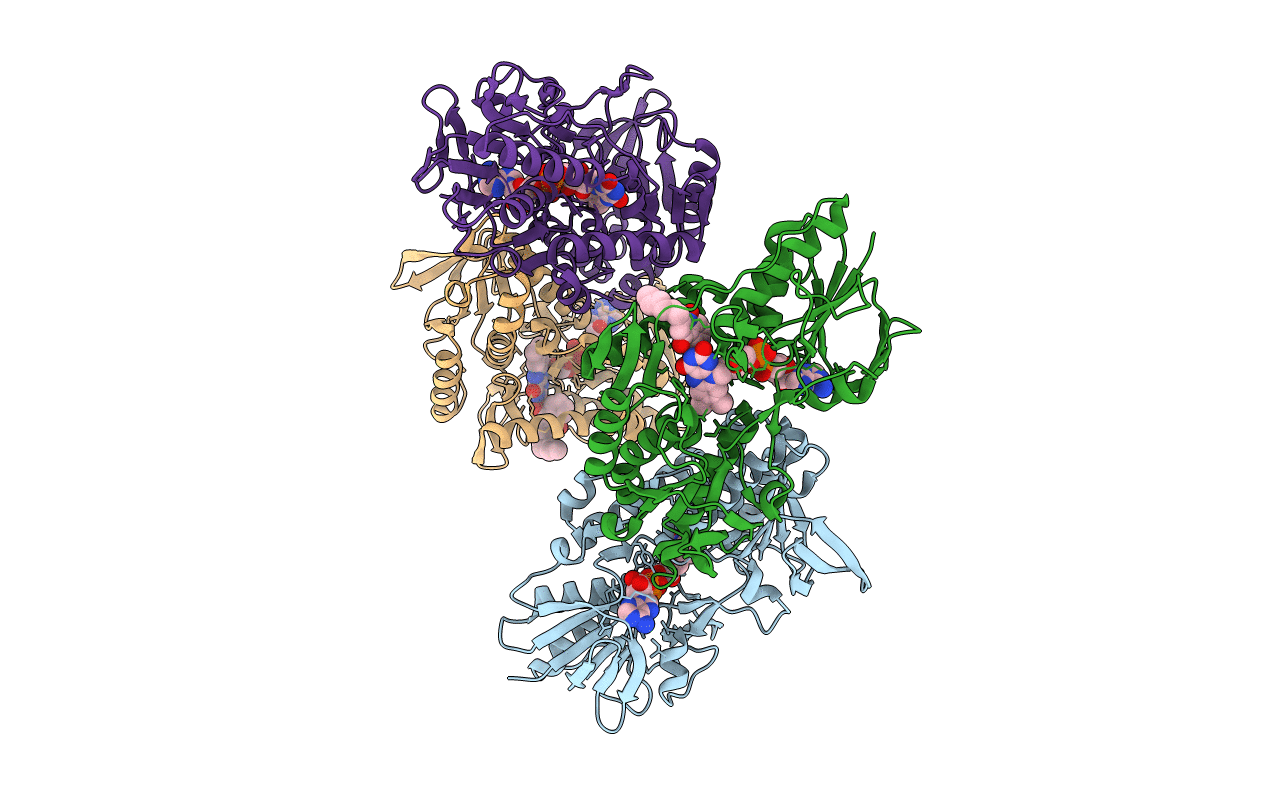
Structure of bacterial type II NADH dehydrogenase from Caldalkalibacillus thermarum complexed with a quinone inhibitor HQNO at 2.8A resolution
Biological Source:
Source Organism(s):
Caldalkalibacillus thermarum TA2.A1 (Taxon ID: 986075)
Expression System(s):
Method Details:
Experimental Method:
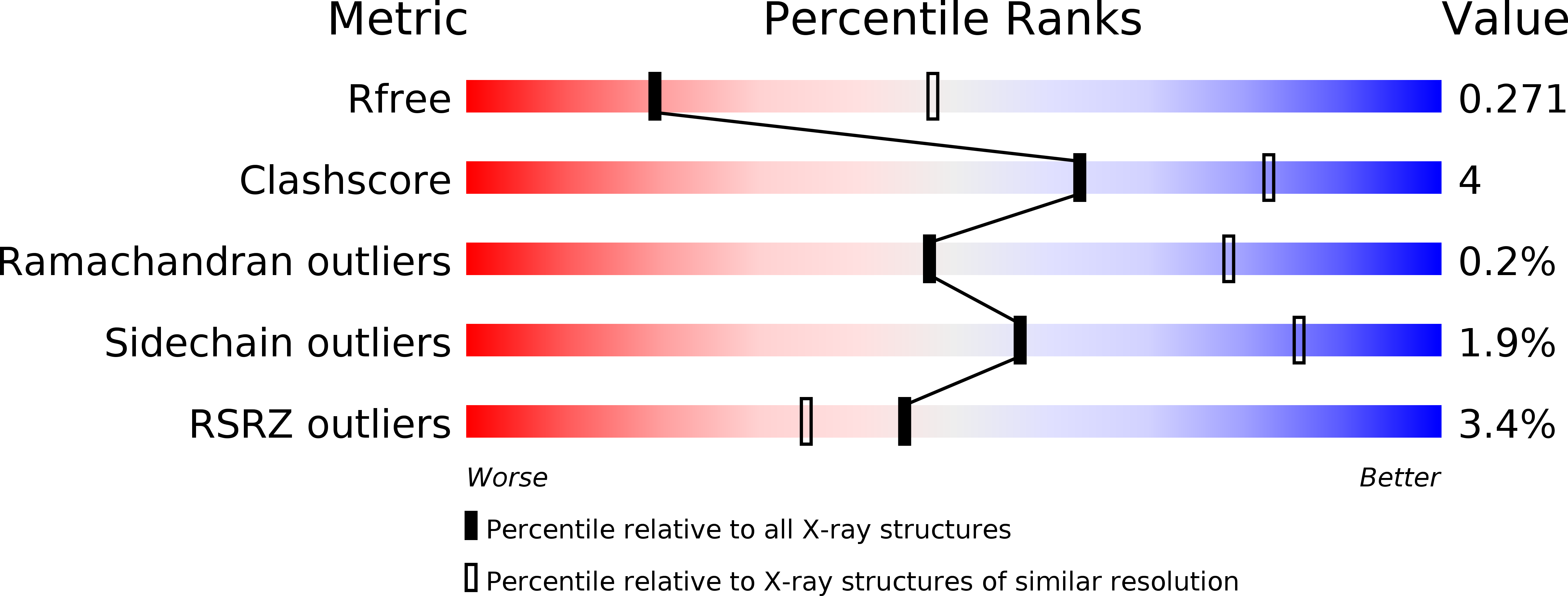
Resolution:
2.80 Å
R-Value Free:
0.26
R-Value Work:
0.22
R-Value Observed:
0.22
Space Group:
P 1 21 1