Deposition Date
2017-10-20
Release Date
2018-10-24
Last Version Date
2024-03-13
Entry Detail
PDB ID:
6BCT
Keywords:
Title:
I-LtrI E184D bound to non-cognate C4 substrate (pre-cleavage complex)
Biological Source:
Source Organism(s):
Leptographium truncatum (Taxon ID: 330483)
Expression System(s):
Method Details:
Experimental Method:
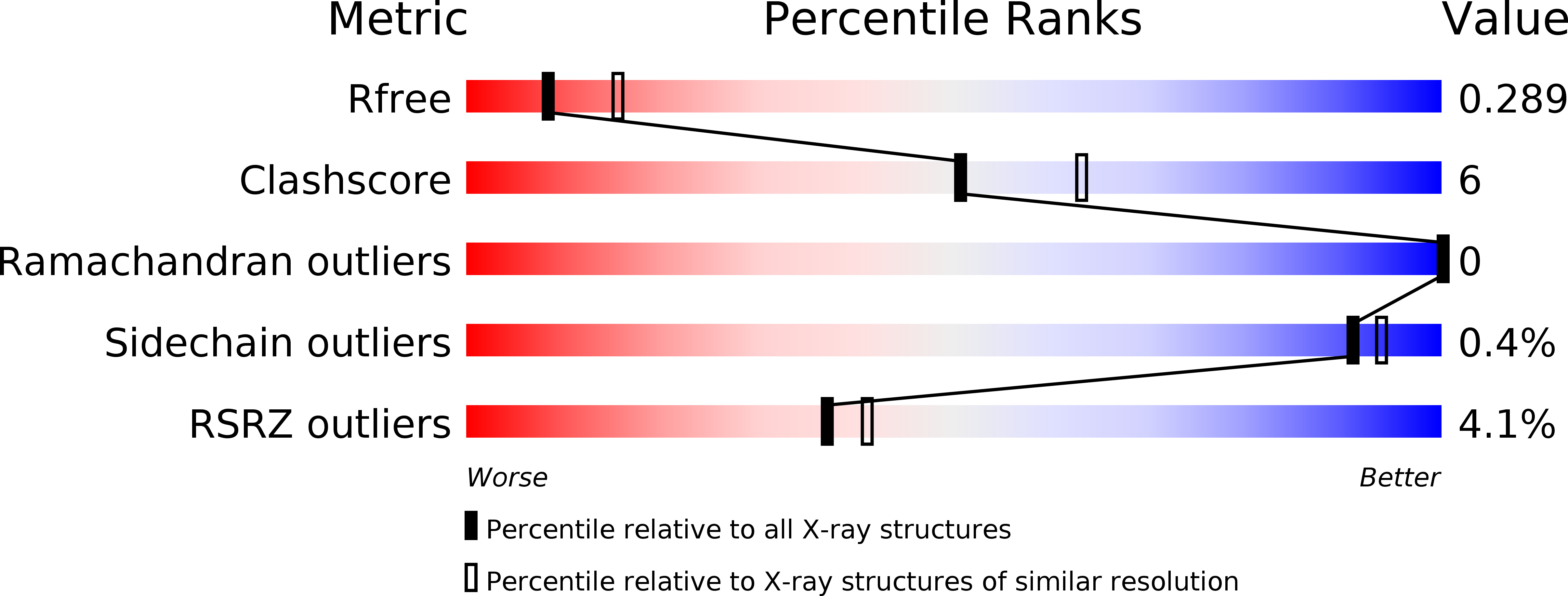
Resolution:
2.73 Å
R-Value Free:
0.28
R-Value Work:
0.22
R-Value Observed:
0.22
Space Group:
C 1 2 1