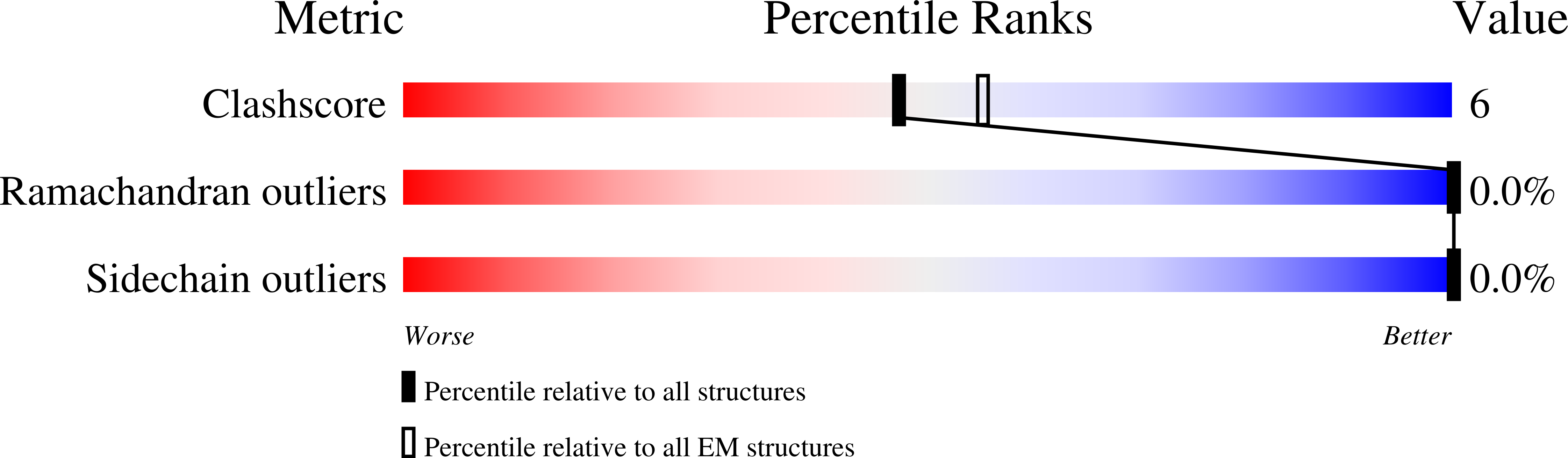
Deposition Date
2017-09-18
Release Date
2017-09-27
Last Version Date
2024-11-06
Entry Detail
PDB ID:
6B1T
Keywords:
Title:
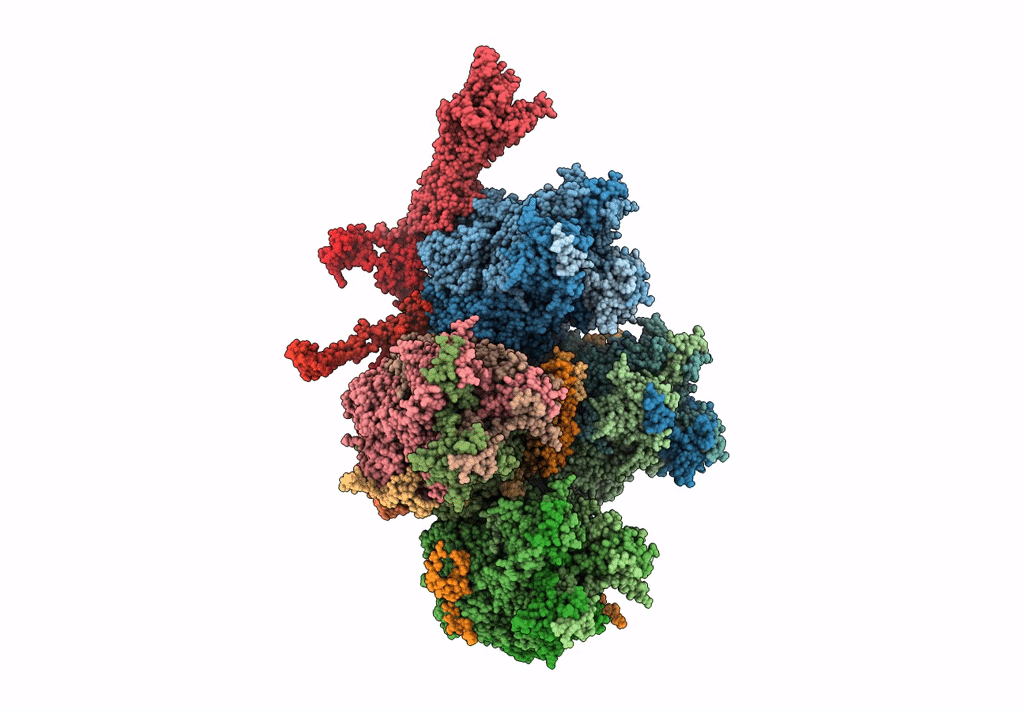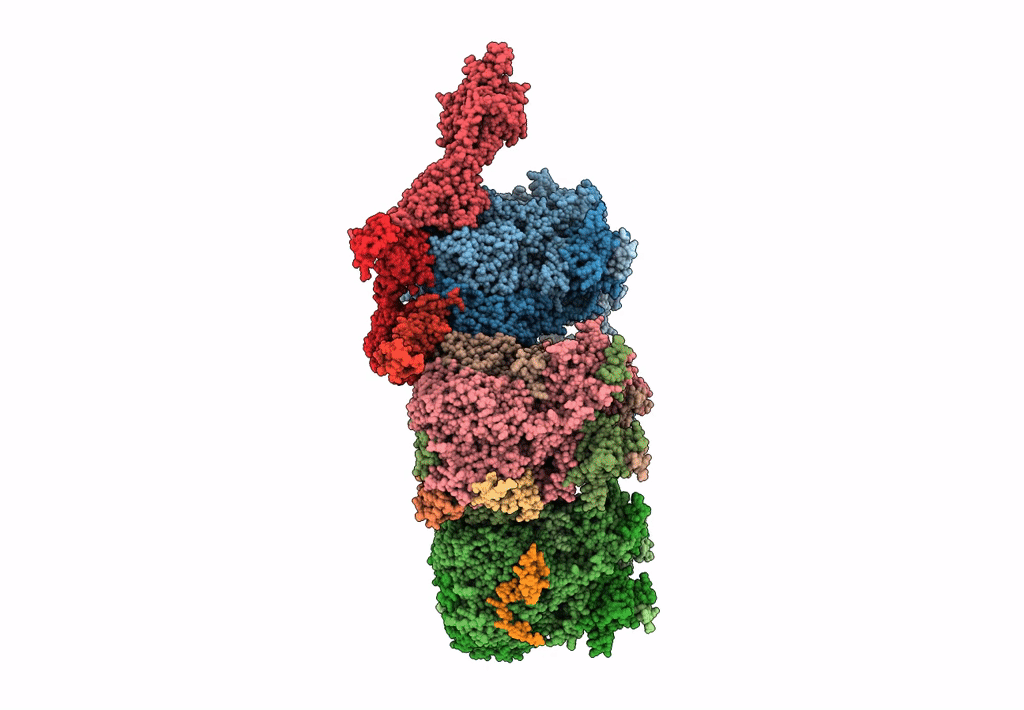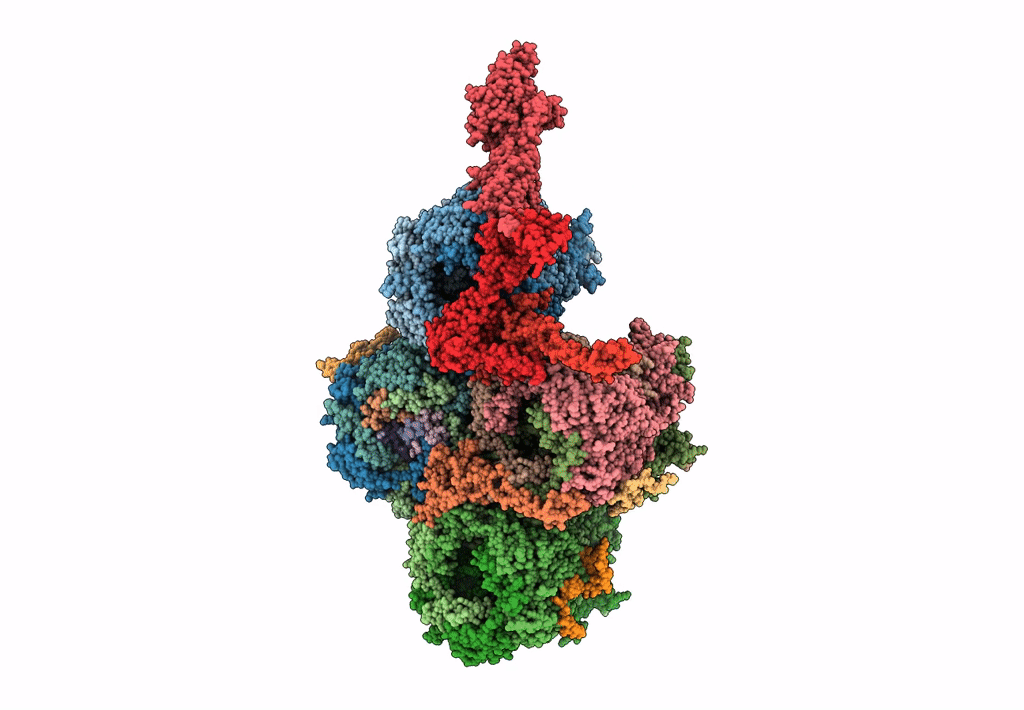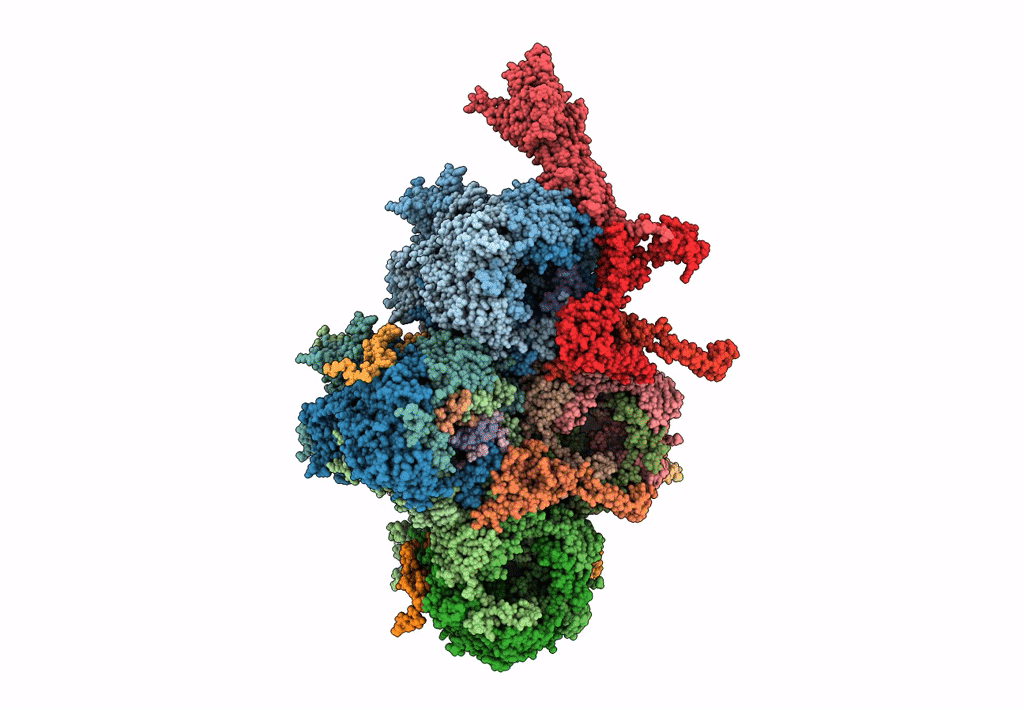
Improved cryoEM structure of human adenovirus type 5 with atomic details of minor proteins VI and VII
Biological Source:
Source Organism(s):
Human adenovirus C serotype 5 (Taxon ID: 28285)
Method Details:
Experimental Method:
Resolution:
3.20 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE