Deposition Date
2017-09-04
Release Date
2018-08-15
Last Version Date
2023-10-04
Entry Detail
PDB ID:
6AVT
Keywords:
Title:
STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING D4TTP AT PH 9.5 WITH CROSS-LINKING TO FIRST BASE TEMPLATE OVERHANG
Biological Source:
Source Organism(s):
Human immunodeficiency virus type 1 group M subtype B (isolate BH10) (Taxon ID: 11678)
Homo sapiens (Taxon ID: 9606)
Homo sapiens (Taxon ID: 9606)
Expression System(s):
Method Details:
Experimental Method:
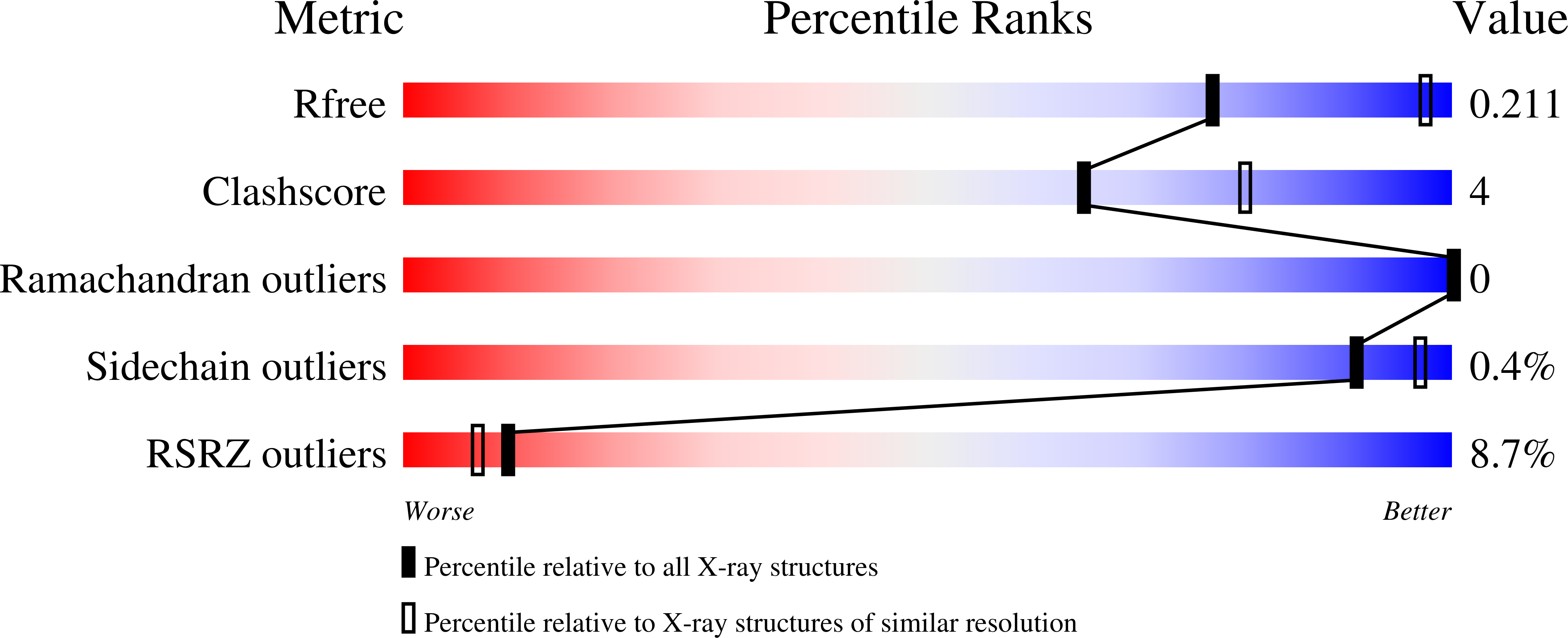
Resolution:
2.60 Å
R-Value Free:
0.21
R-Value Work:
0.17
R-Value Observed:
0.17
Space Group:
P 1 21 1