Deposition Date
2018-09-03
Release Date
2019-02-13
Last Version Date
2024-11-13
Entry Detail
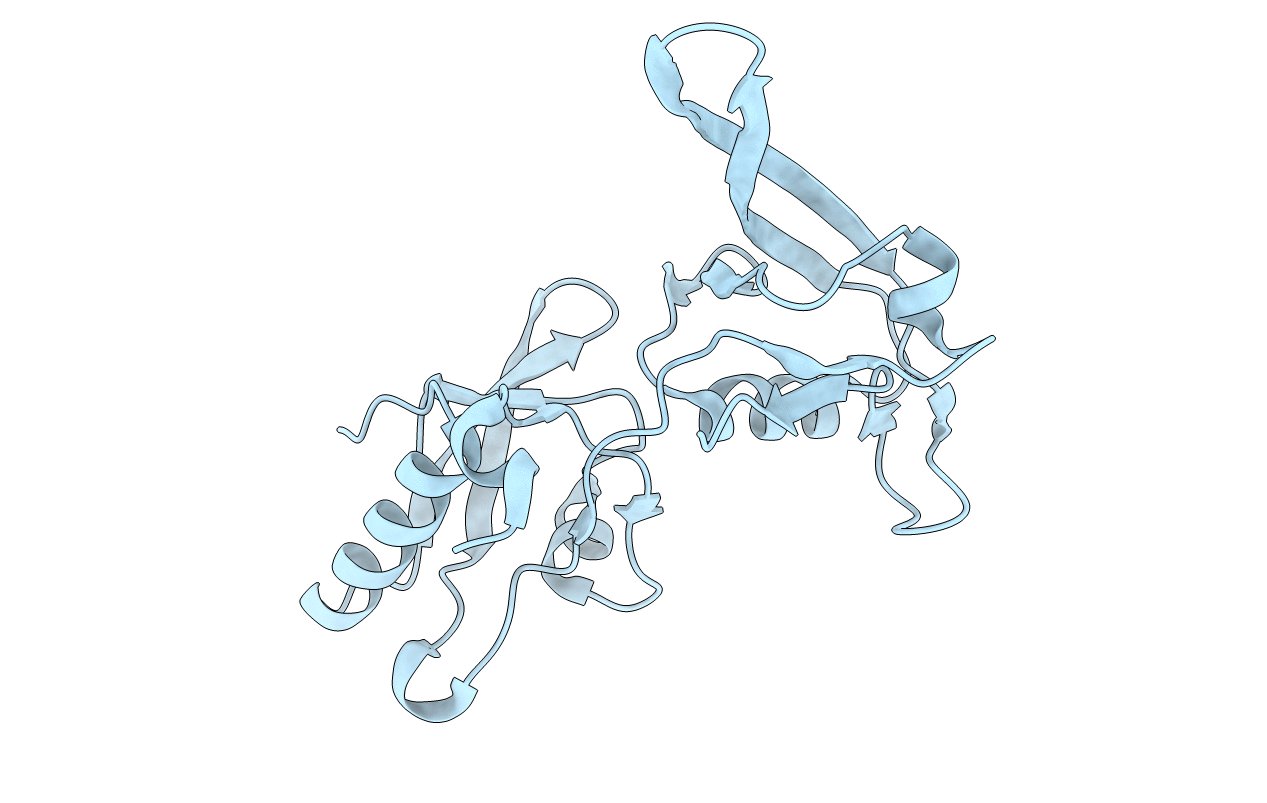
PDB ID:
6AKQ
Keywords:
Title:
The PDZ tandem fragment of A. aeolicus S2P homolog with the PA12 tag inserted between the residues 263 and 267
Biological Source:
Source Organism:
Aquifex aeolicus VF5 (Taxon ID: 224324)
Host Organism:
Method Details:
Experimental Method:
Resolution:
1.90 Å
R-Value Free:
0.25
R-Value Work:
0.20
R-Value Observed:
0.20
Space Group:
P 21 21 21


