Deposition Date
2018-07-26
Release Date
2018-08-08
Last Version Date
2024-10-23
Entry Detail
PDB ID:
6ACD
Keywords:
Title:
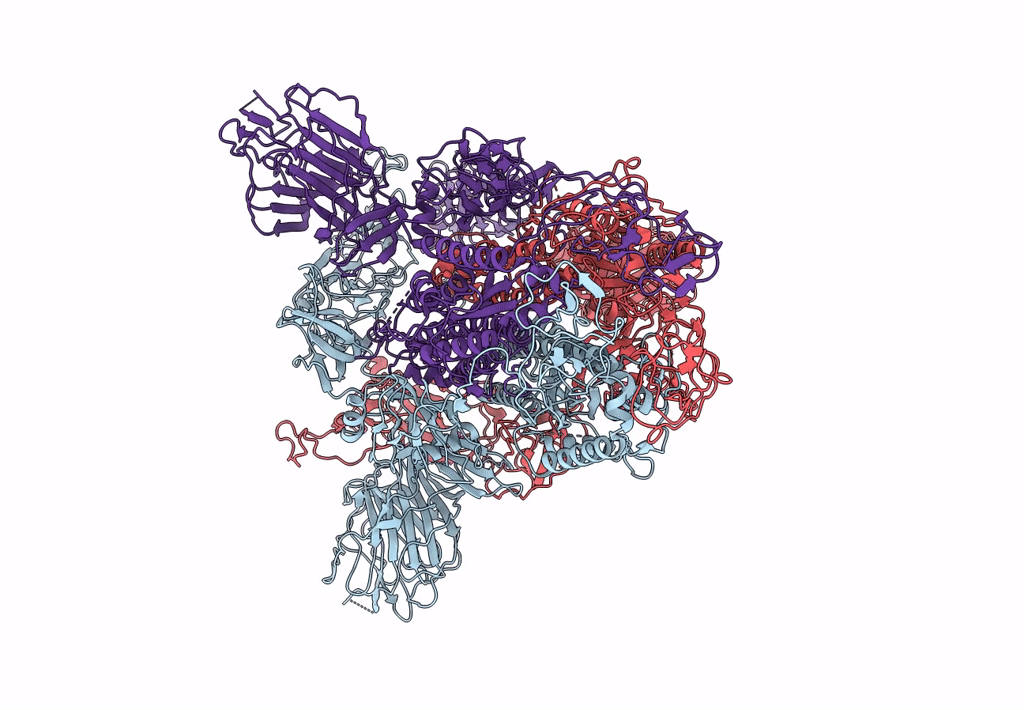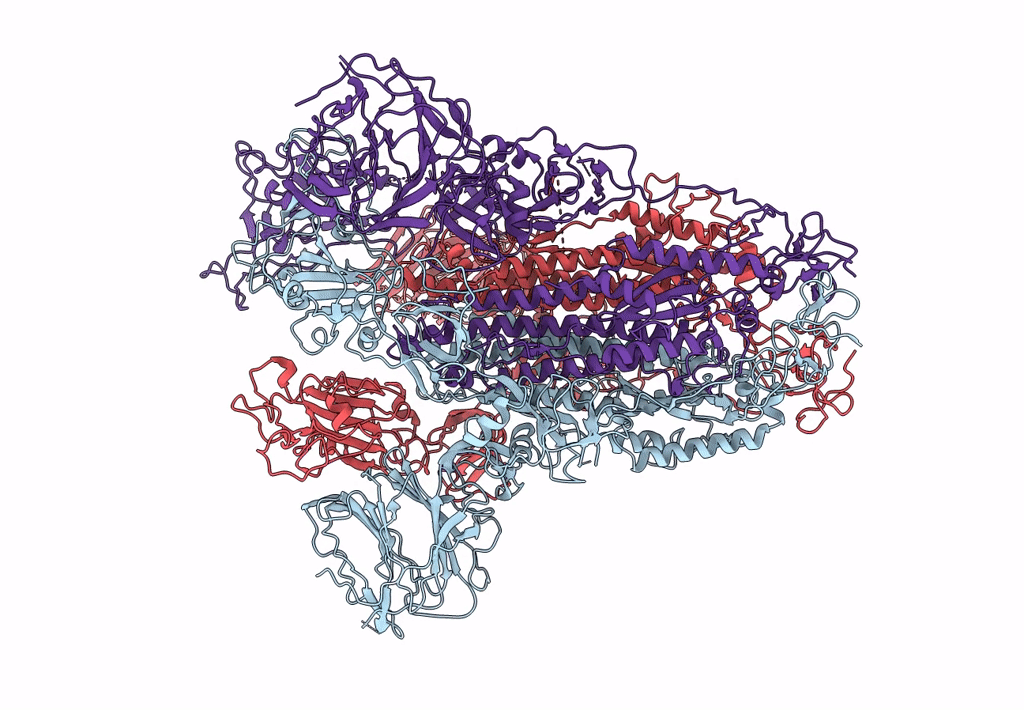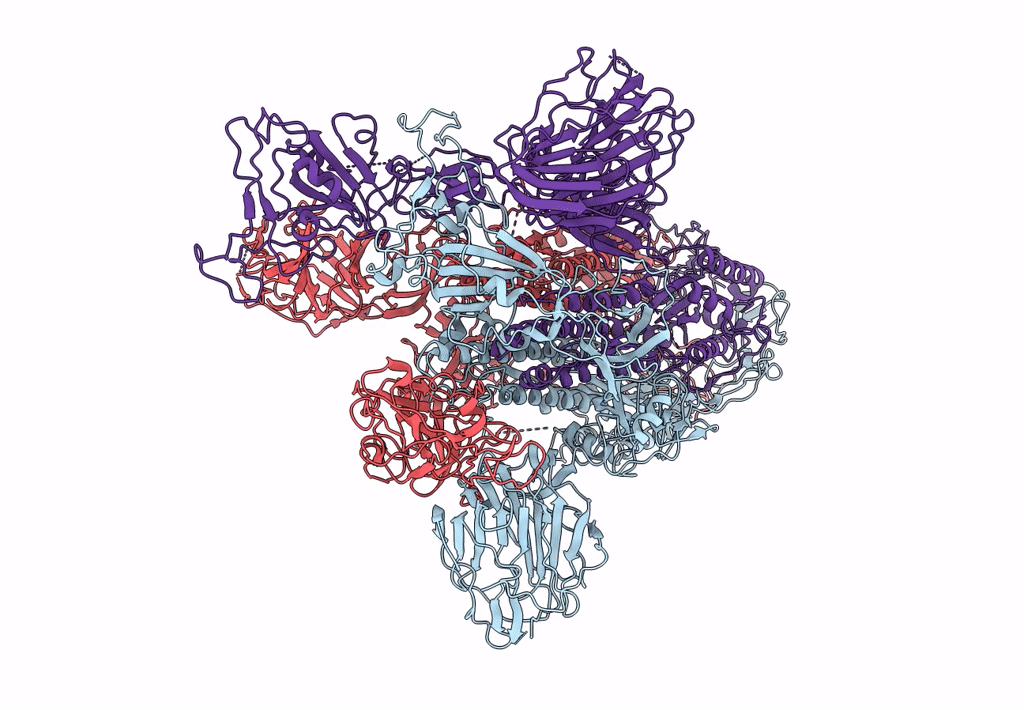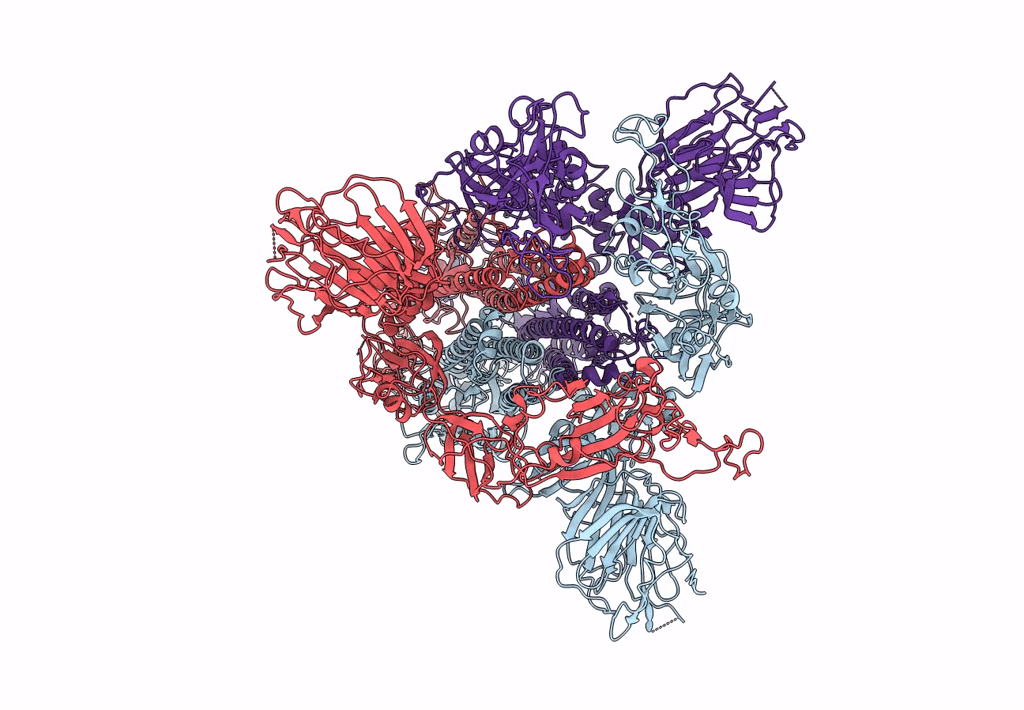
Trypsin-cleaved and low pH-treated SARS-CoV spike glycoprotein and ACE2 complex, ACE2-free conformation with one RBD in up conformation
Biological Source:
Source Organism(s):
Human SARS coronavirus (Taxon ID: 227859)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
3.90 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE


