Deposition Date
2018-06-28
Release Date
2019-07-03
Last Version Date
2024-10-23
Entry Detail
PDB ID:
6A6H
Keywords:
Title:
Crystal Structure of Swine Major Histocompatibility Complex Class I SLA-2*040202 For 2.3 Angstrom
Biological Source:
Source Organism:
Sus scrofa (Taxon ID: 9823)
Foot-and-mouth disease virus - type A (Taxon ID: 12111)
Foot-and-mouth disease virus - type A (Taxon ID: 12111)
Host Organism:
Method Details:
Experimental Method:
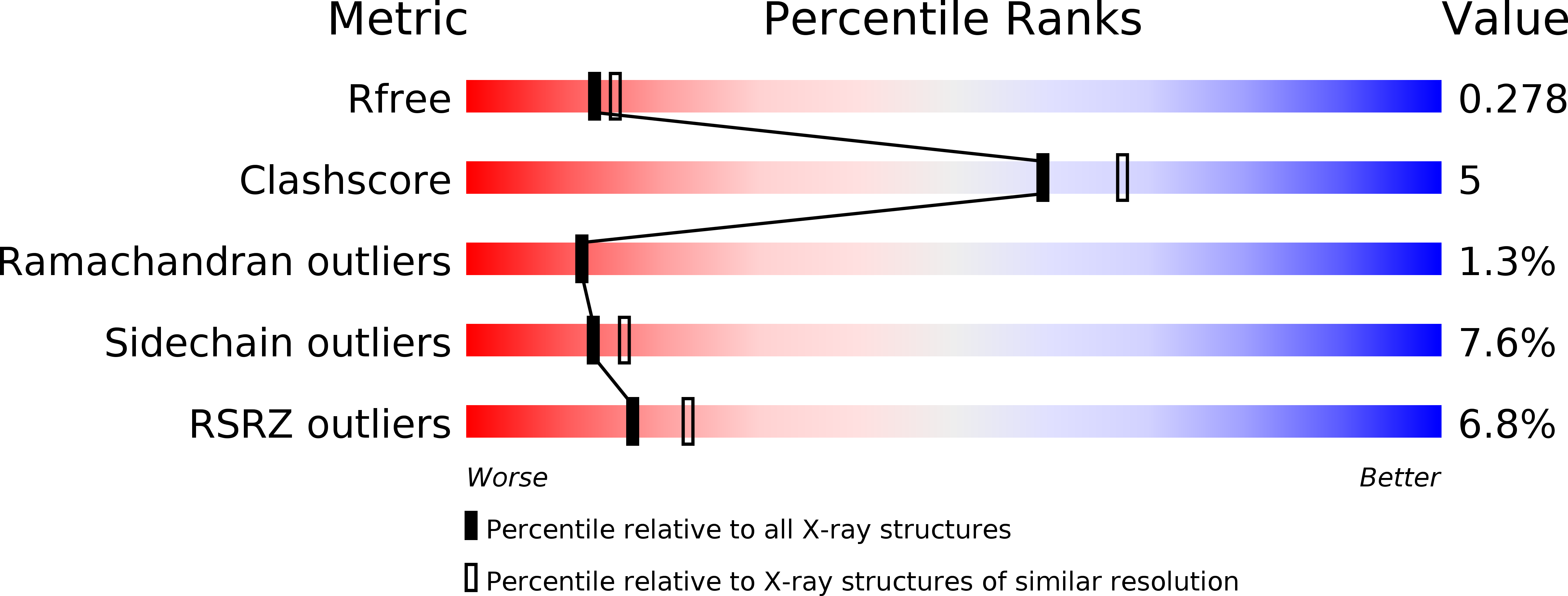
Resolution:
2.31 Å
R-Value Free:
0.27
R-Value Work:
0.19
R-Value Observed:
0.20
Space Group:
P 32 2 1