Deposition Date
2017-08-13
Release Date
2018-01-24
Last Version Date
2023-11-22
Entry Detail
PDB ID:
5Y6Q
Keywords:
Title:
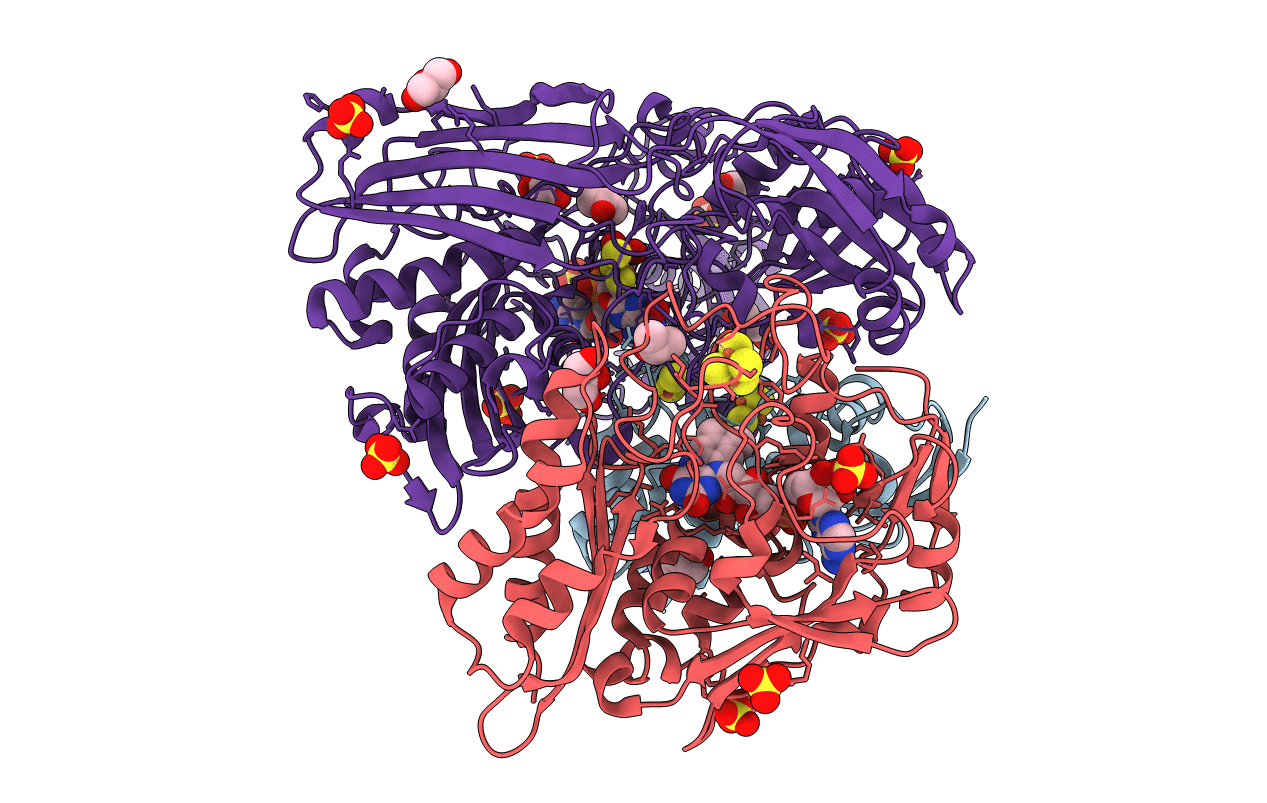
Crystal structure of an aldehyde oxidase from Methylobacillus sp. KY4400
Biological Source:
Source Organism:
Methylobacillus sp. KY4400 (Taxon ID: 194289)
Host Organism:
Method Details:
Experimental Method:
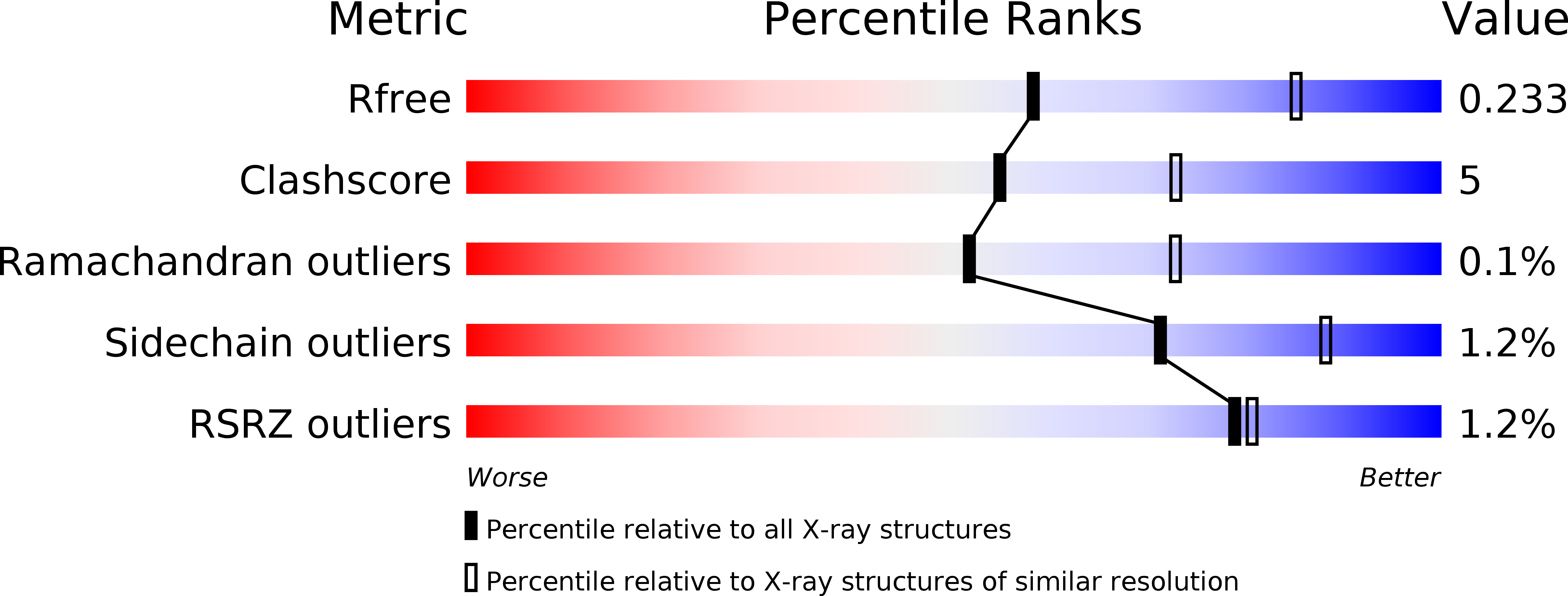
Resolution:
2.50 Å
R-Value Free:
0.23
R-Value Work:
0.17
R-Value Observed:
0.18
Space Group:
P 41 21 2