Deposition Date
2017-05-24
Release Date
2017-07-19
Last Version Date
2023-11-22
Entry Detail
PDB ID:
5XNX
Keywords:
Title:
Crystallographic structure of the enzymatically active N-terminal domain of the Rel protein from Mycobacterium tuberculosis
Biological Source:
Source Organism:
Host Organism:
Method Details:
Experimental Method:
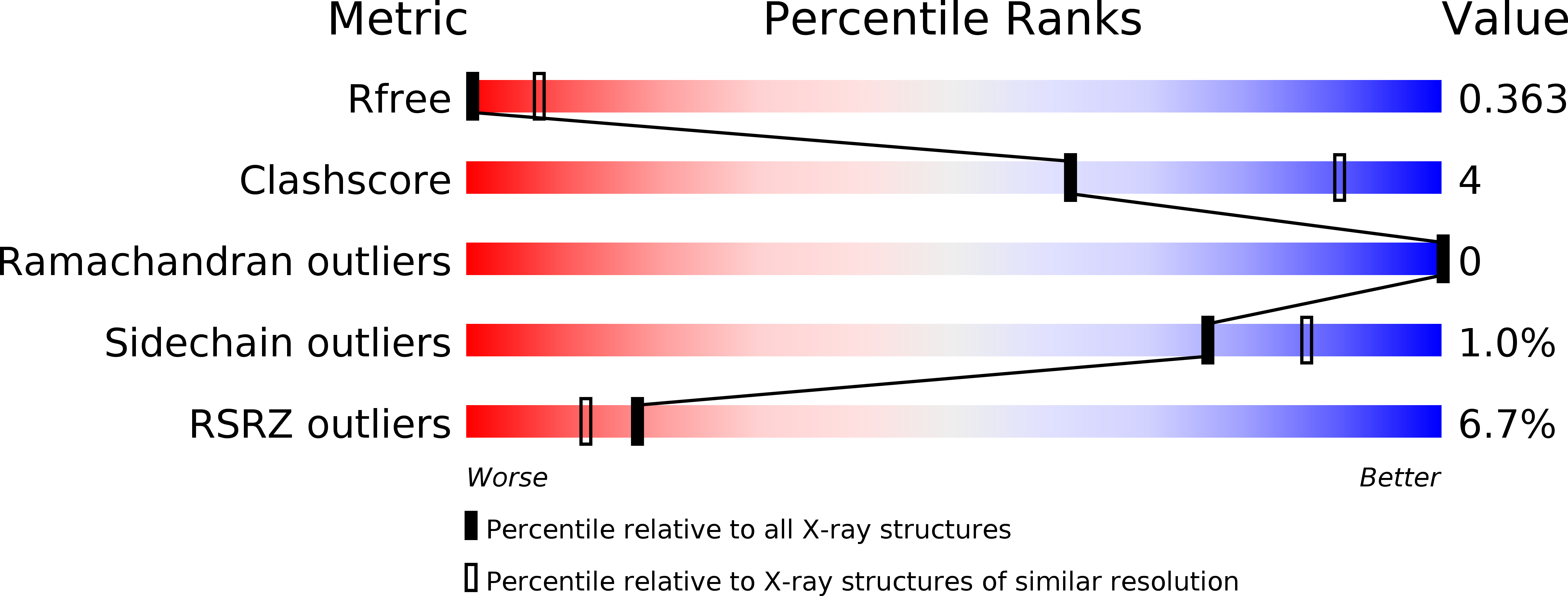
Resolution:
3.70 Å
R-Value Free:
0.36
R-Value Work:
0.35
R-Value Observed:
0.35
Space Group:
P 32