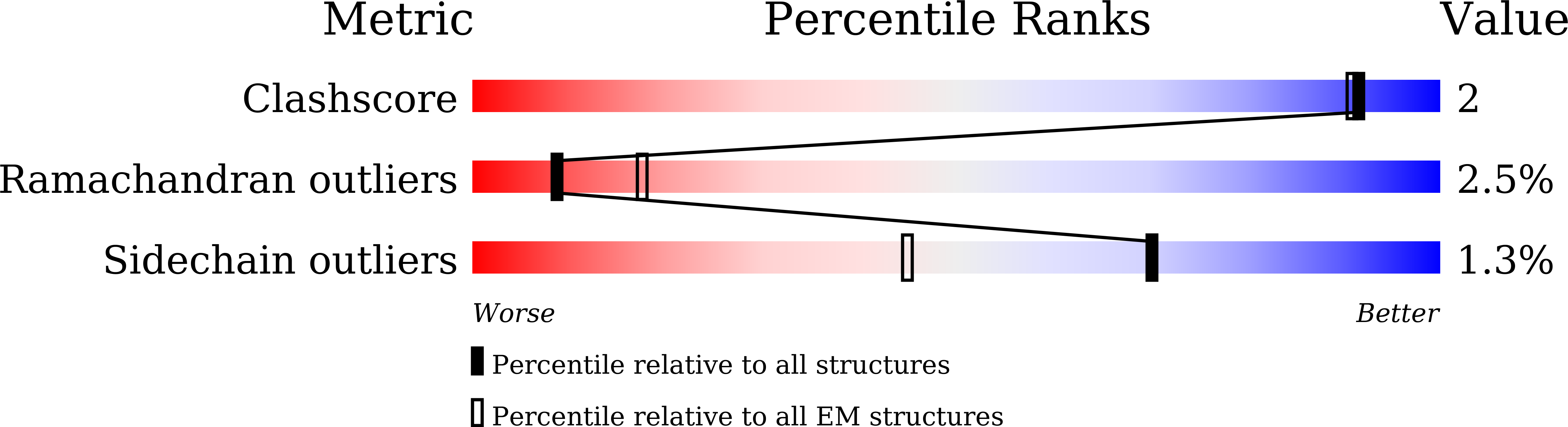Deposition Date
2017-05-15
Release Date
2017-08-09
Last Version Date
2024-03-27
Entry Detail
PDB ID:
5XMI
Keywords:
Title:
Cryo-EM Structure of the ATP-bound VPS4 mutant-E233Q hexamer (masked)
Biological Source:
Source Organism(s):
Expression System(s):
Method Details:
Experimental Method:
Resolution:
3.90 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE