Deposition Date
2017-07-25
Release Date
2017-09-06
Last Version Date
2023-10-04
Entry Detail
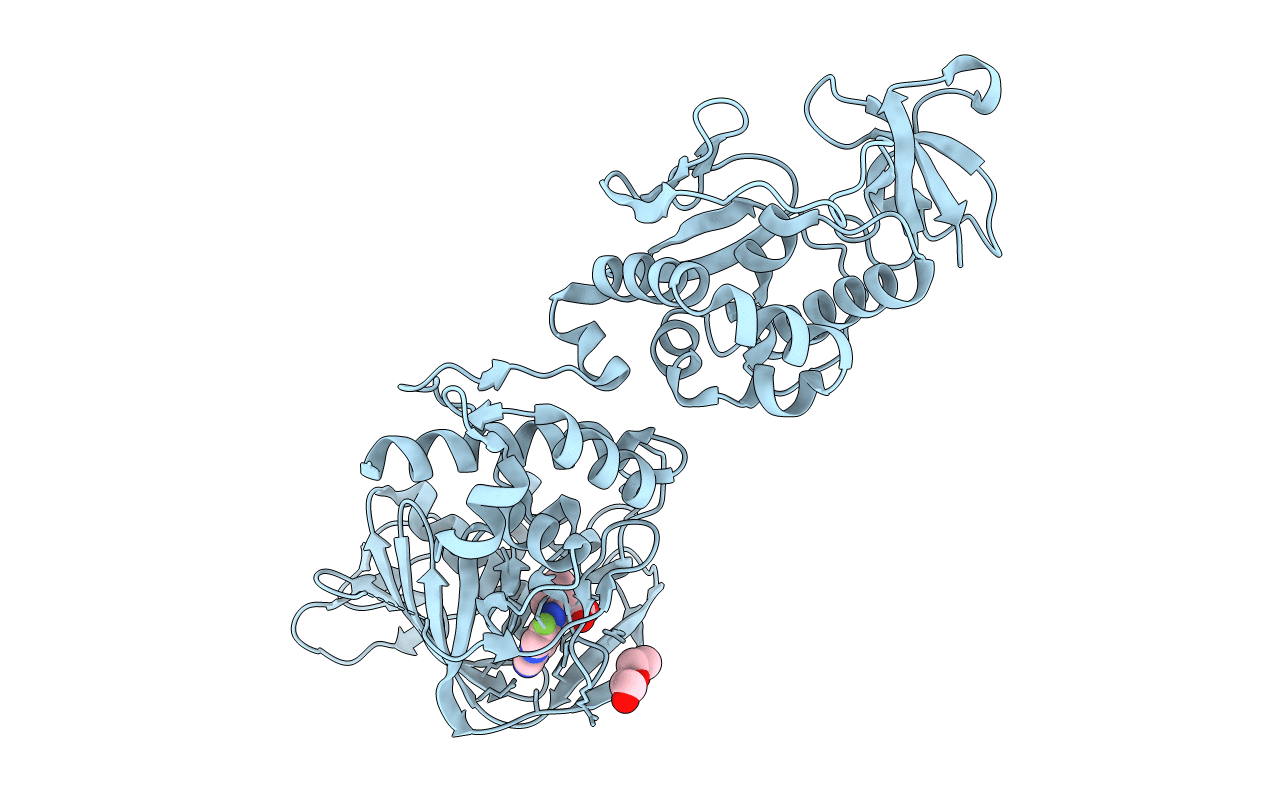
PDB ID:
5WL0
Keywords:
Title:
Co-crystal structure of Influenza A H3N2 PB2 (241-741) bound to VX-787
Biological Source:
Source Organism(s):
Expression System(s):
Method Details:
Experimental Method:
Resolution:
2.40 Å
R-Value Free:
0.25
R-Value Work:
0.20
R-Value Observed:
0.20
Space Group:
C 1 2 1


