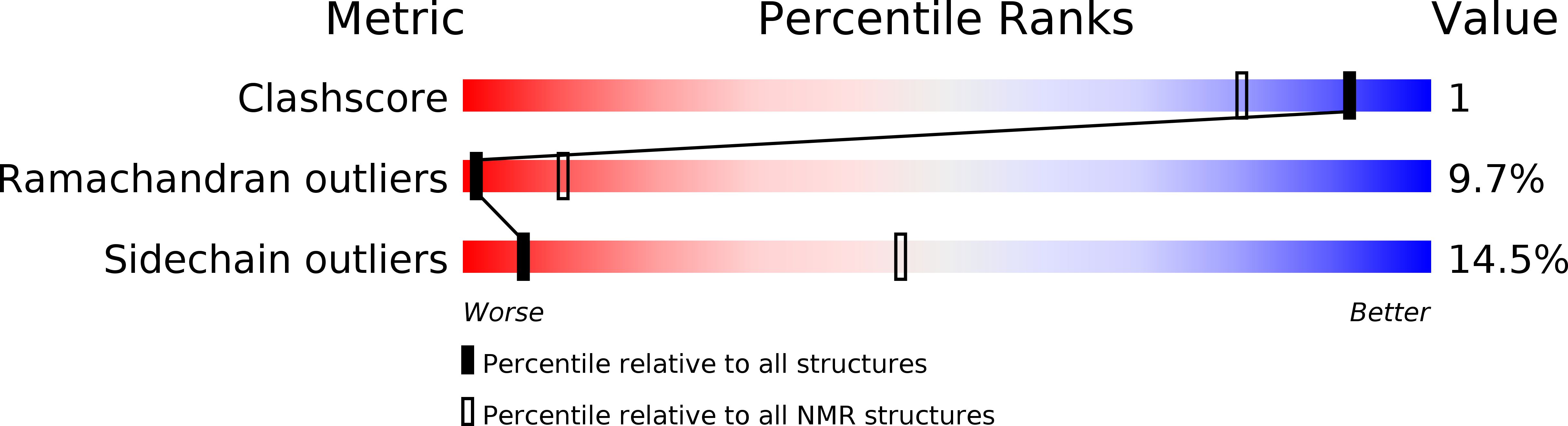
Deposition Date
2017-06-08
Release Date
2017-09-27
Last Version Date
2024-05-15
Entry Detail
PDB ID:
5W3N
Keywords:
Title:
Molecular structure of FUS low sequence complexity domain protein fibrils
Biological Source:
Source Organism:
Homo sapiens (Taxon ID: 9606)
Host Organism:
Method Details:
Experimental Method:
Conformers Calculated:
4928
Conformers Submitted:
20
Selection Criteria:
structures with no violations, lowest energy, and derived from one of 44 independent calculations