Deposition Date
2017-06-01
Release Date
2017-12-13
Last Version Date
2024-11-20
Entry Detail
PDB ID:
5W11
Keywords:
Title:
Biochemical and structural insights into the catalytic mechanism of thermostable cellobiohydrolase Cel7A from industrially relevant fungus Myceliophthora thermophila
Biological Source:
Source Organism(s):
Myceliophthora thermophila (Taxon ID: 78579)
Method Details:
Experimental Method:
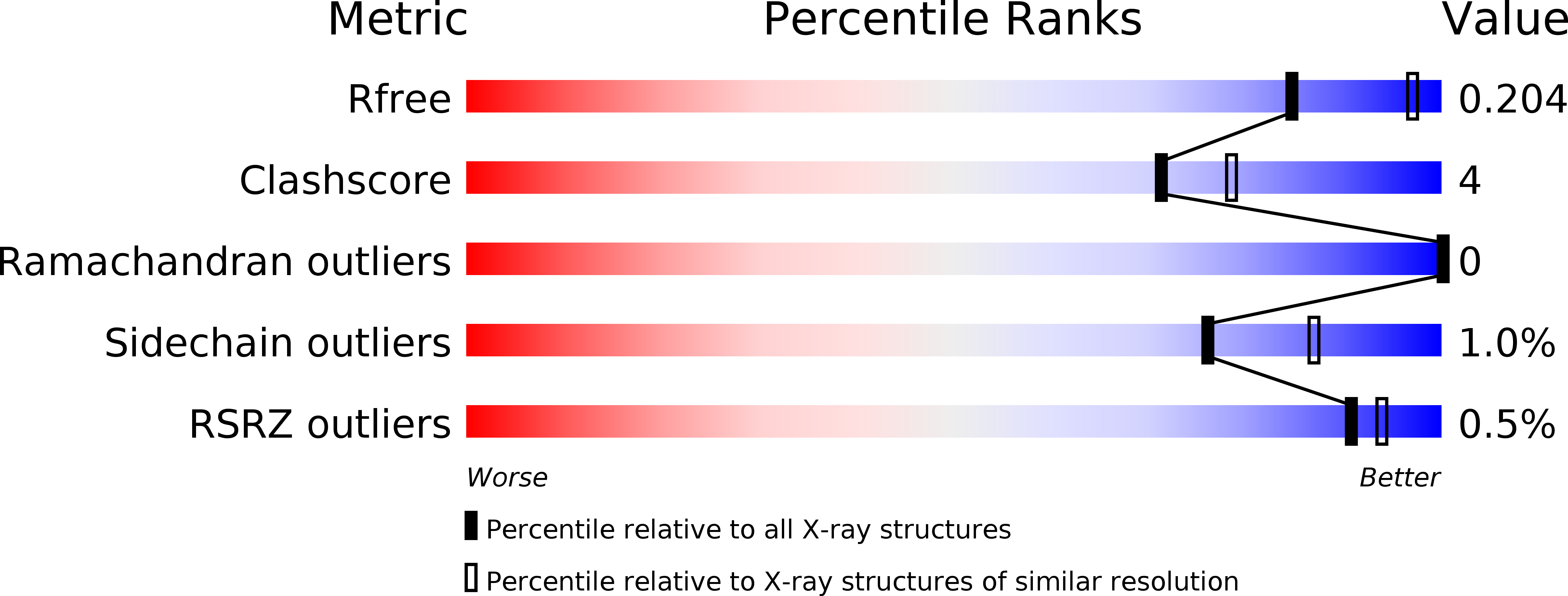
Resolution:
2.31 Å
R-Value Free:
0.20
R-Value Work:
0.13
R-Value Observed:
0.14
Space Group:
P 1 21 1