Deposition Date
2017-05-18
Release Date
2017-12-06
Last Version Date
2024-10-16
Entry Detail
PDB ID:
5VU2
Keywords:
Title:
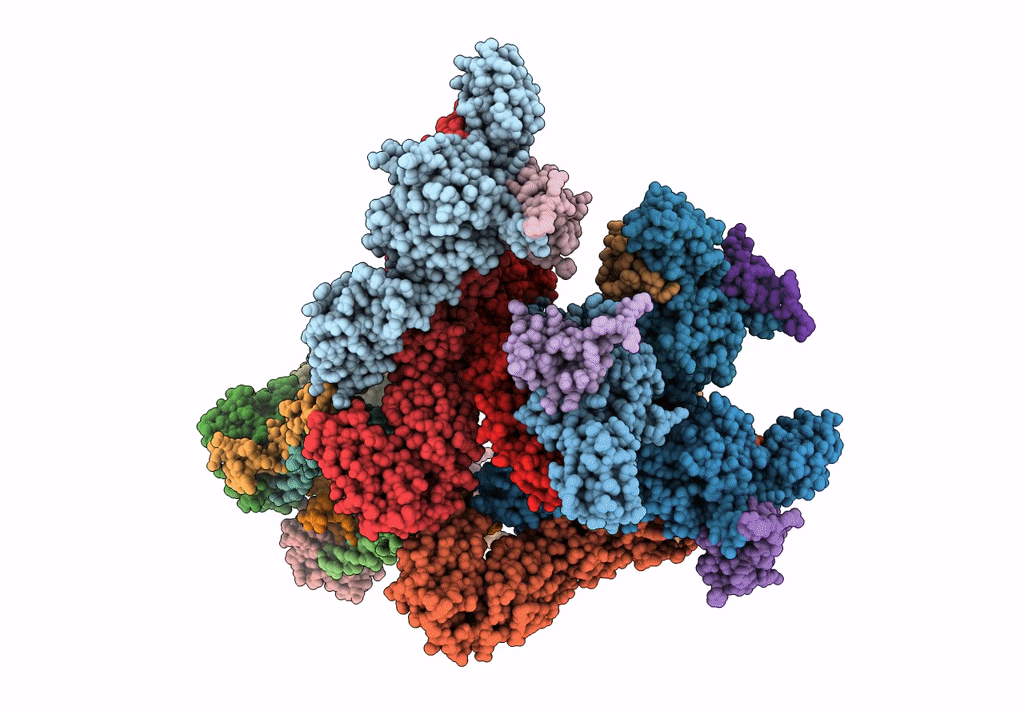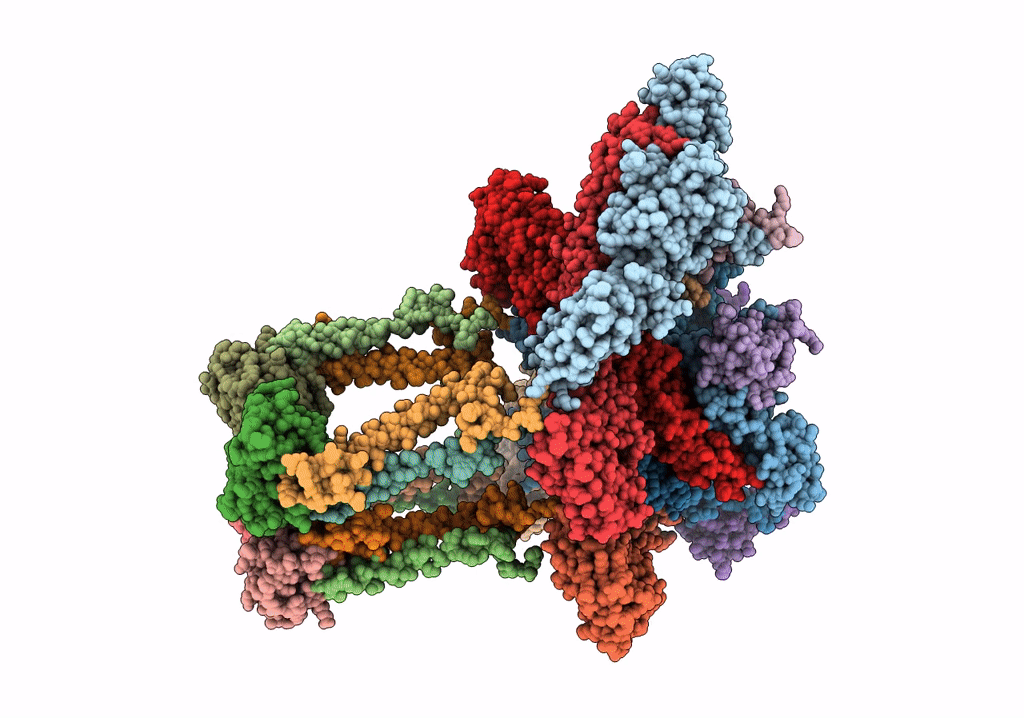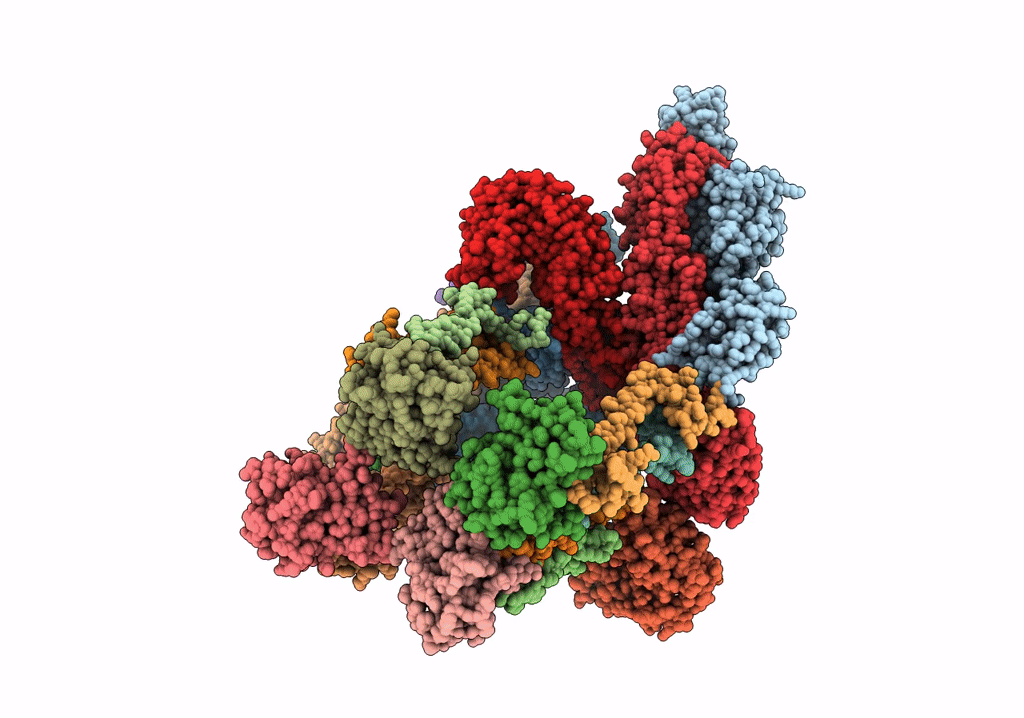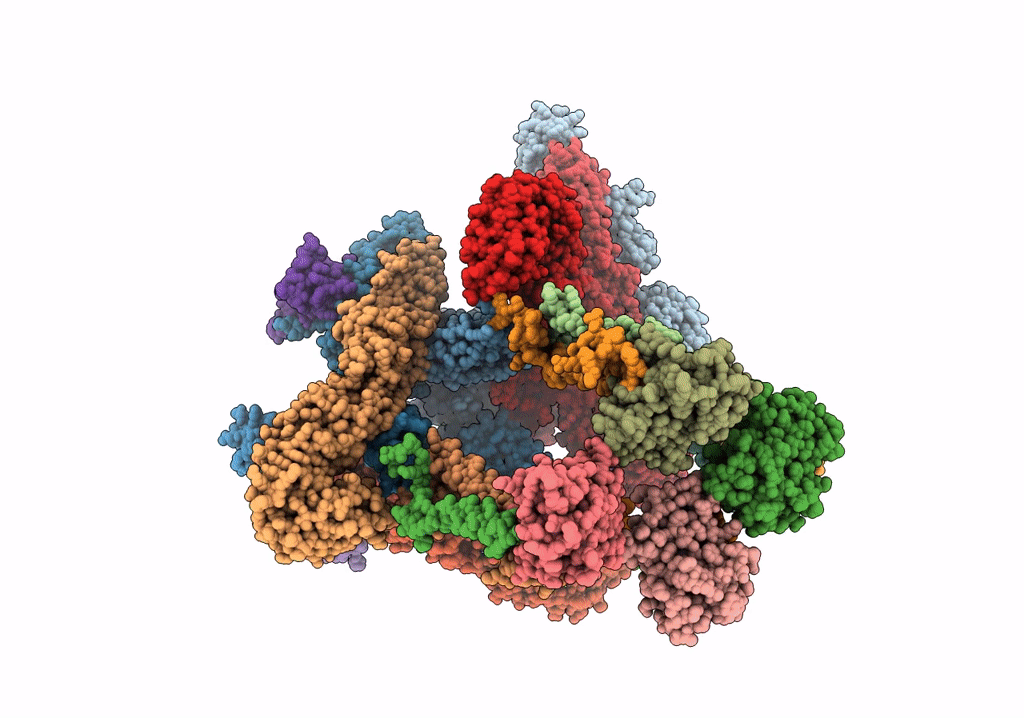
Electron cryo-microscopy of "immature" Chikungunya VLP
Biological Source:
Source Organism(s):
Chikungunya virus strain Senegal 37997 (Taxon ID: 371095)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
6.80 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE


