Deposition Date
2017-03-02
Release Date
2018-02-14
Last Version Date
2024-03-20
Entry Detail
PDB ID:
5V1D
Keywords:
Title:
Complex structure of the bovine PERK luminal domain and its substrate peptide
Biological Source:
Source Organism(s):
Bos taurus (Taxon ID: 9913)
synthetic construct (Taxon ID: 32630)
synthetic construct (Taxon ID: 32630)
Expression System(s):
Method Details:
Experimental Method:
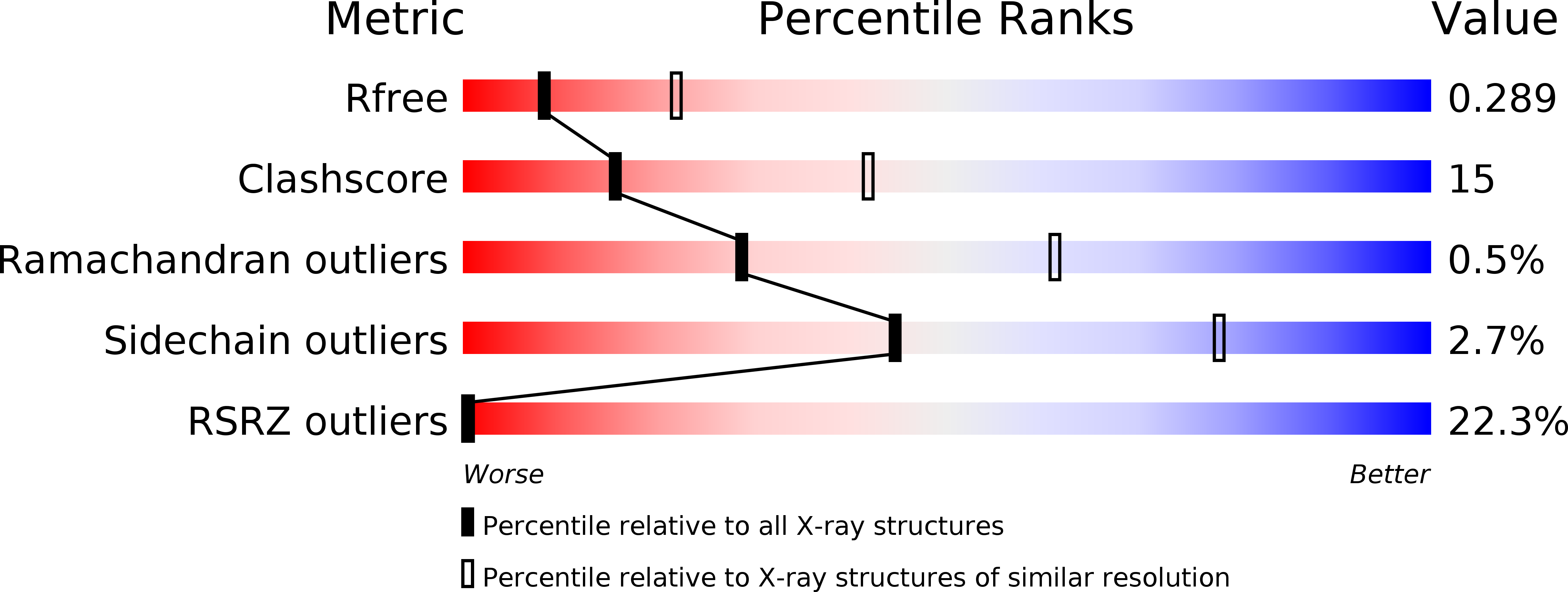
Resolution:
2.80 Å
R-Value Free:
0.29
R-Value Work:
0.24
R-Value Observed:
0.24
Space Group:
P 1 21 1