Deposition Date
2017-01-17
Release Date
2017-02-01
Last Version Date
2024-05-01
Entry Detail
PDB ID:
5UJ5
Keywords:
Title:
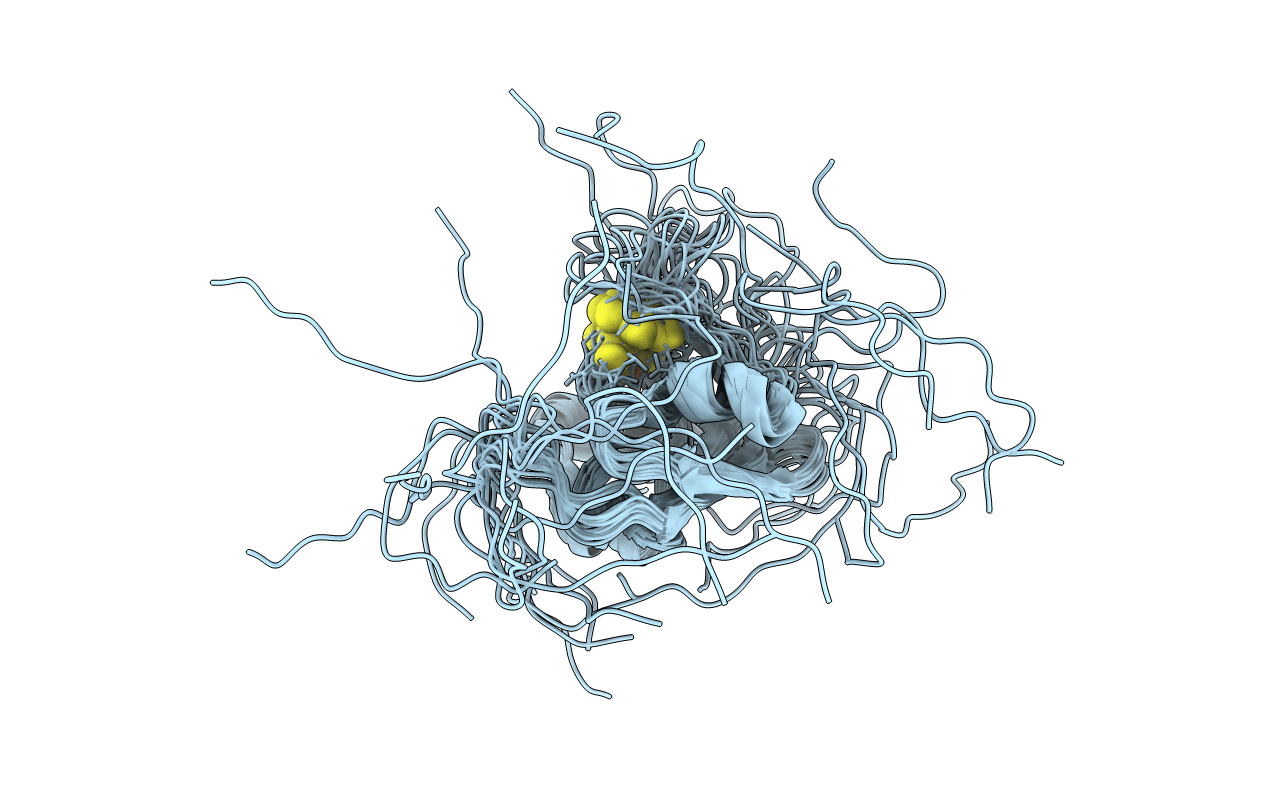
Solution structure of the oxidized iron-sulfur protein adrenodoxin from Encephalitozoon cuniculi. Seattle Structural Genomics Center for Infectious Disease target EncuA.00705.a
Biological Source:
Source Organism(s):
Encephalitozoon cuniculi (Taxon ID: 6035)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
100
Conformers Submitted:
20
Selection Criteria:
target function