Deposition Date
2016-10-05
Release Date
2017-10-18
Last Version Date
2023-10-04
Entry Detail
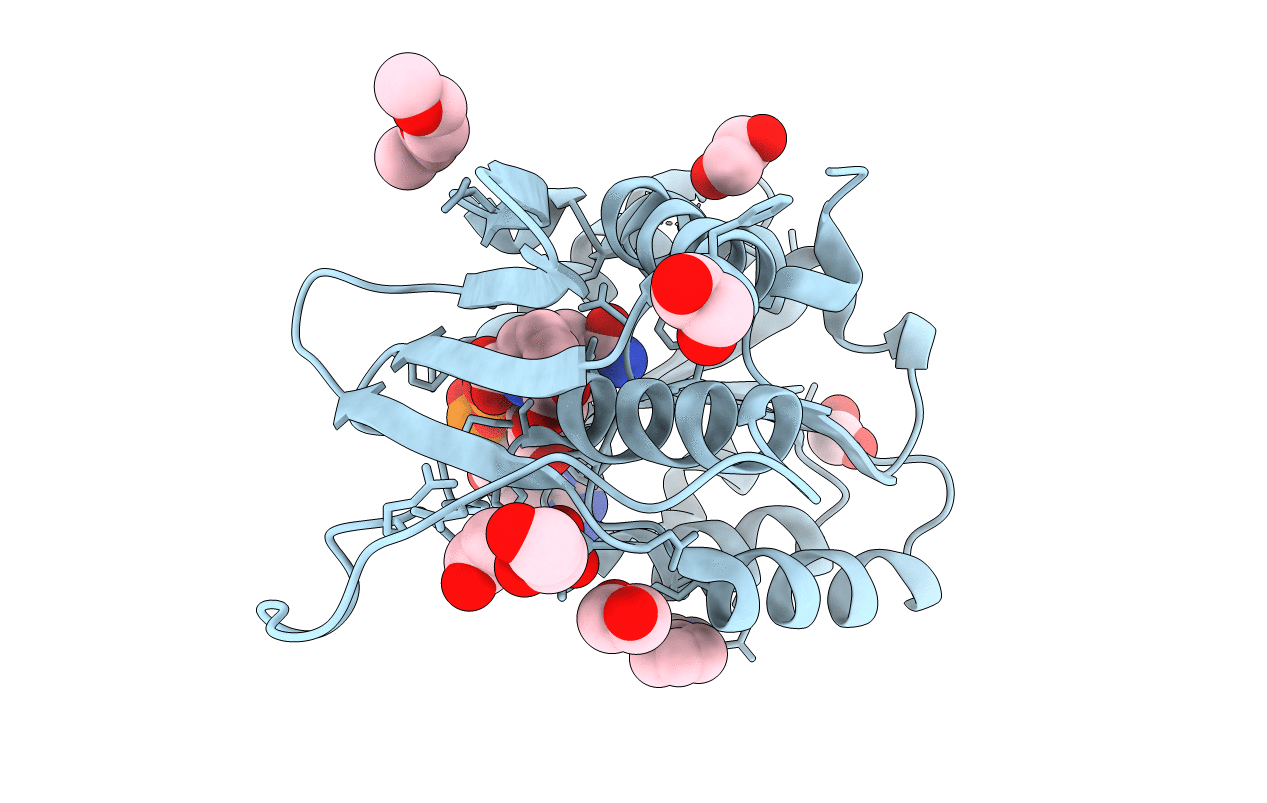
PDB ID:
5TJY
Keywords:
Title:
Structure of 4-Hydroxy-tetrahydrodipicolinate Reductase from Mycobacterium tuberculosis with 2,6 Pyridine Dicarboxylic Acid and NADH
Biological Source:
Source Organism:
Host Organism:
Method Details:
Experimental Method:
Resolution:
2.40 Å
R-Value Free:
0.19
R-Value Work:
0.14
R-Value Observed:
0.14
Space Group:
P 62 2 2


