Deposition Date
2016-09-13
Release Date
2016-12-07
Last Version Date
2024-10-30
Entry Detail
Biological Source:
Source Organism:
Escherichia coli O157:H7 (Taxon ID: 83334)
Escherichia coli O45:K1 (strain S88 / ExPEC) (Taxon ID: 585035)
Escherichia coli O45:K1 (strain S88 / ExPEC) (Taxon ID: 585035)
Host Organism:
Method Details:
Experimental Method:
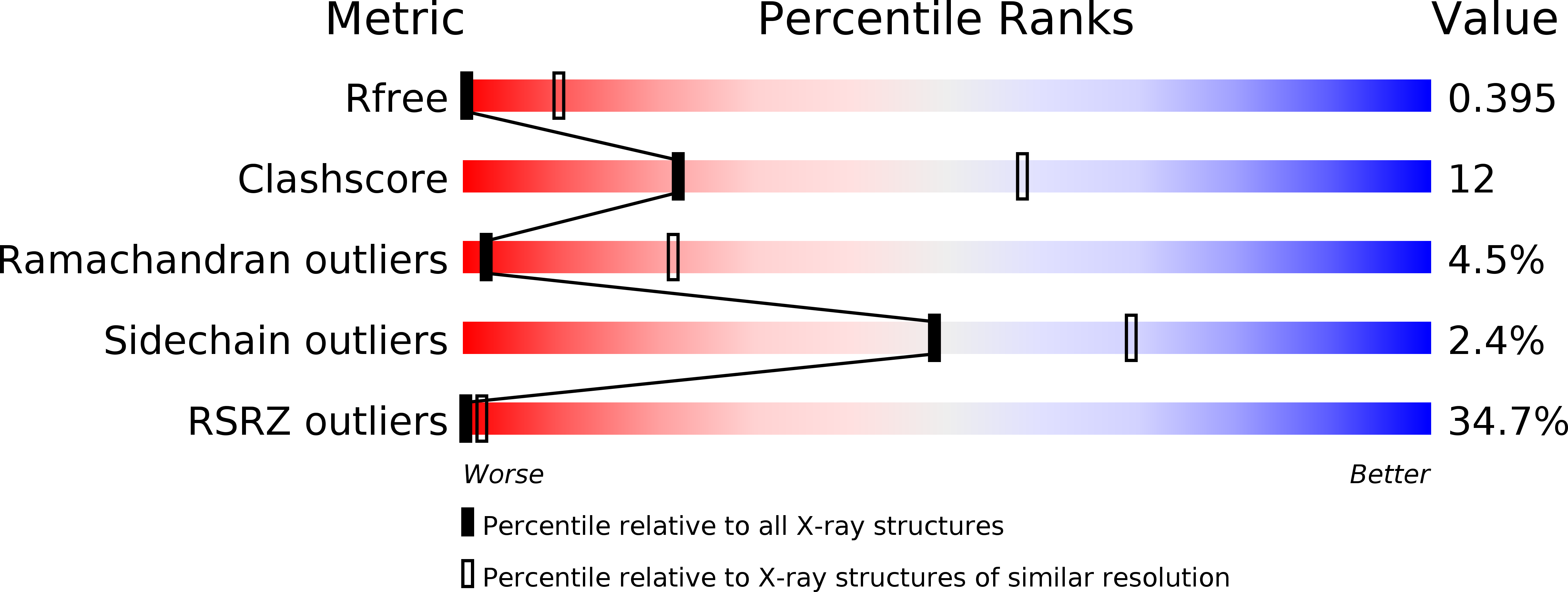
Resolution:
7.00 Å
R-Value Free:
0.39
R-Value Work:
0.33
R-Value Observed:
0.33
Space Group:
C 1 2 1