Deposition Date
2016-09-13
Release Date
2017-06-07
Last Version Date
2024-10-23
Entry Detail
PDB ID:
5TBY
Keywords:
Title:
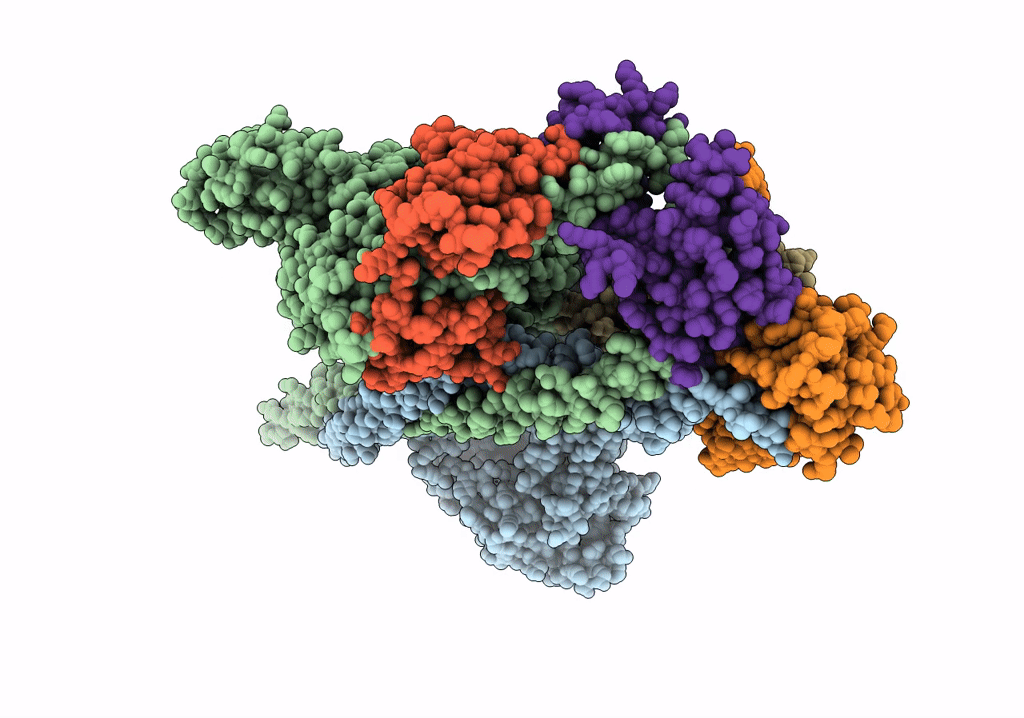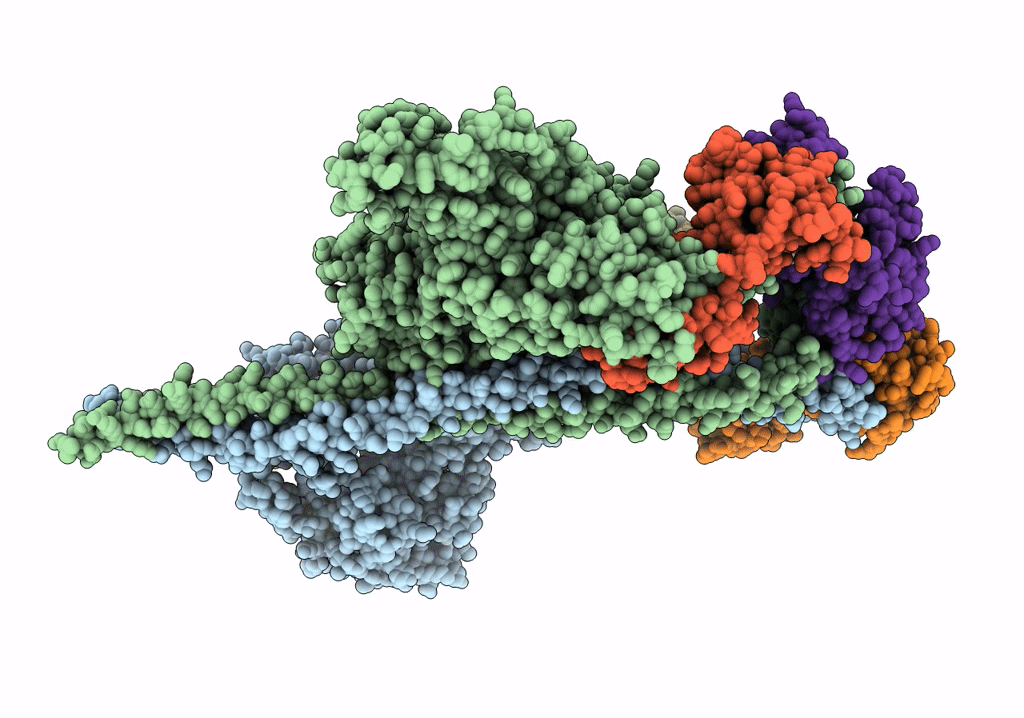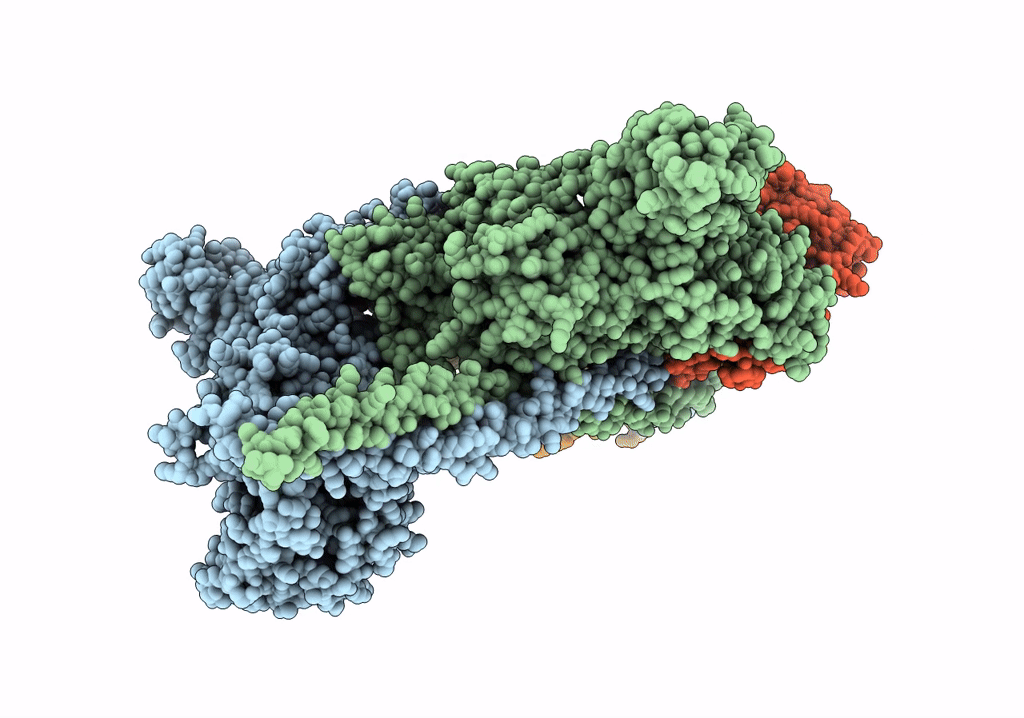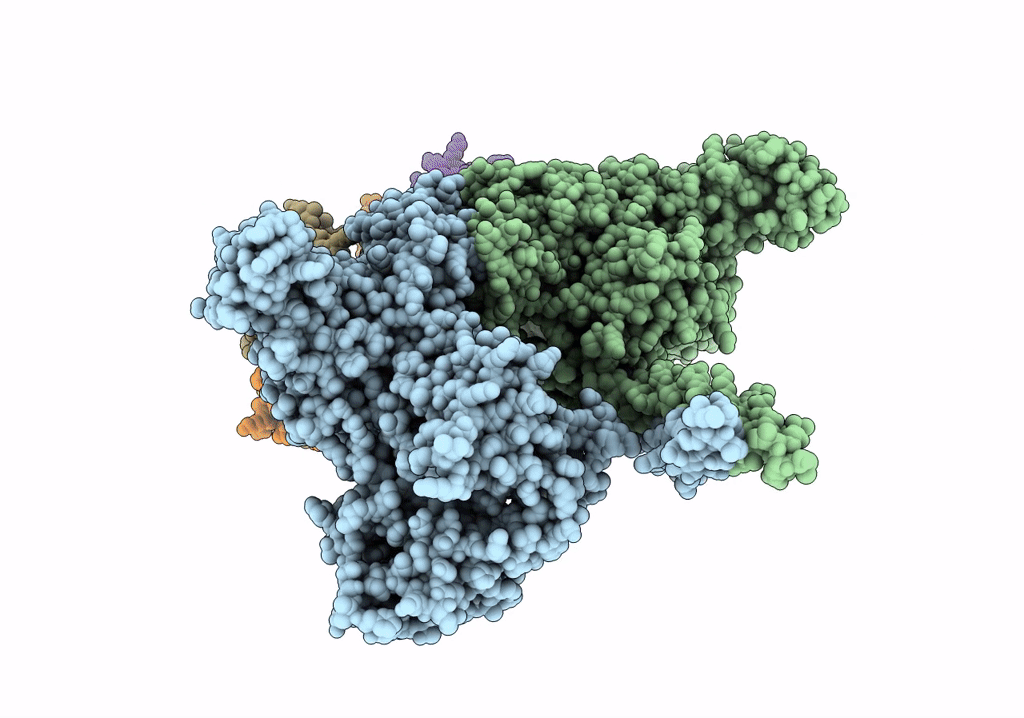
HUMAN BETA CARDIAC HEAVY MEROMYOSIN INTERACTING-HEADS MOTIF OBTAINED BY HOMOLOGY MODELING (USING SWISS-MODEL) OF HUMAN SEQUENCE FROM APHONOPELMA HOMOLOGY MODEL (PDB-3JBH), RIGIDLY FITTED TO HUMAN BETA-CARDIAC NEGATIVELY STAINED THICK FILAMENT 3D-RECONSTRUCTION (EMD-2240)
Biological Source:
Source Organism(s):
Homo sapiens (Taxon ID: 9606)
Method Details:
Experimental Method:
Resolution:
20.00 Å
Aggregation State:
FILAMENT
Reconstruction Method:
SINGLE PARTICLE