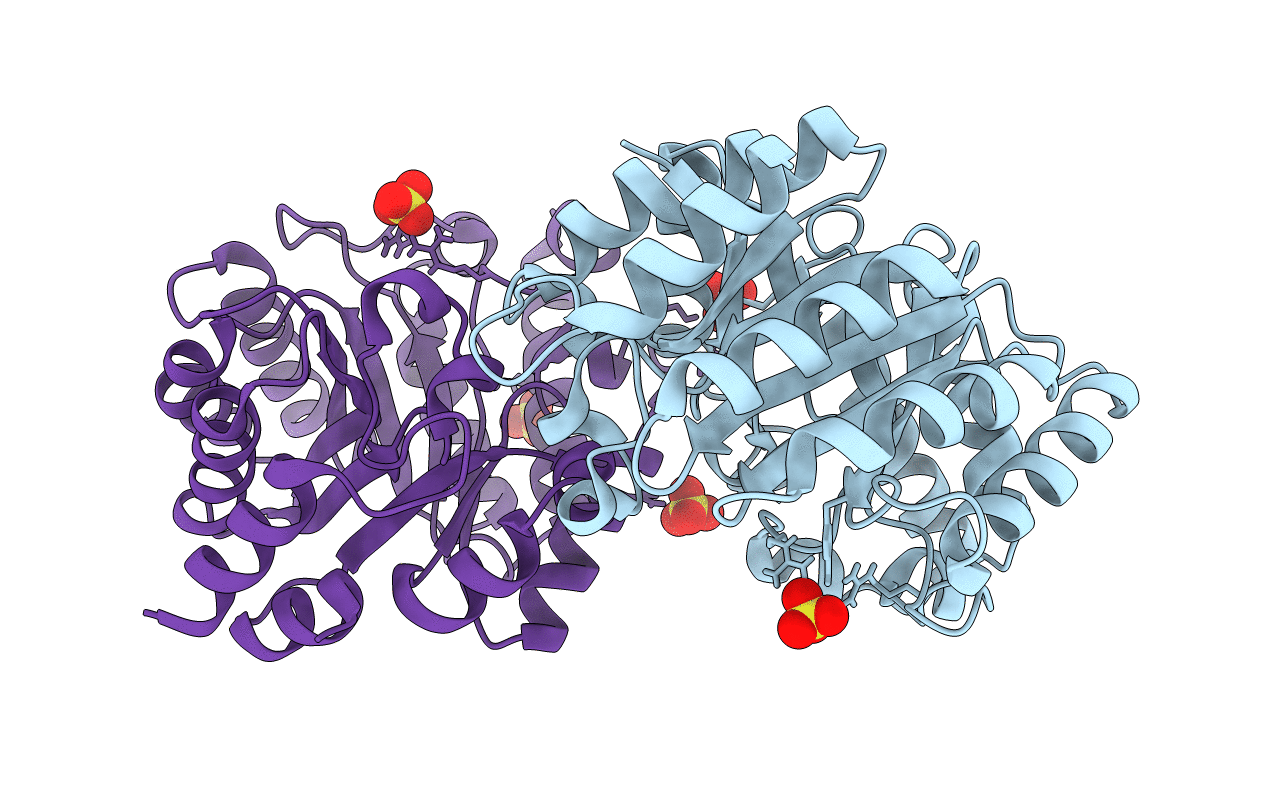
Deposition Date
2020-03-19
Release Date
2020-07-22
Last Version Date
2024-03-06
Entry Detail
PDB ID:
5RG6
Keywords:
Title:
Crystal Structure of Kemp Eliminase HG3.7 in unbound state, 277K
Biological Source:
Source Organism(s):
Thermoascus aurantiacus (Taxon ID: 5087)
Expression System(s):
Method Details:
Experimental Method:
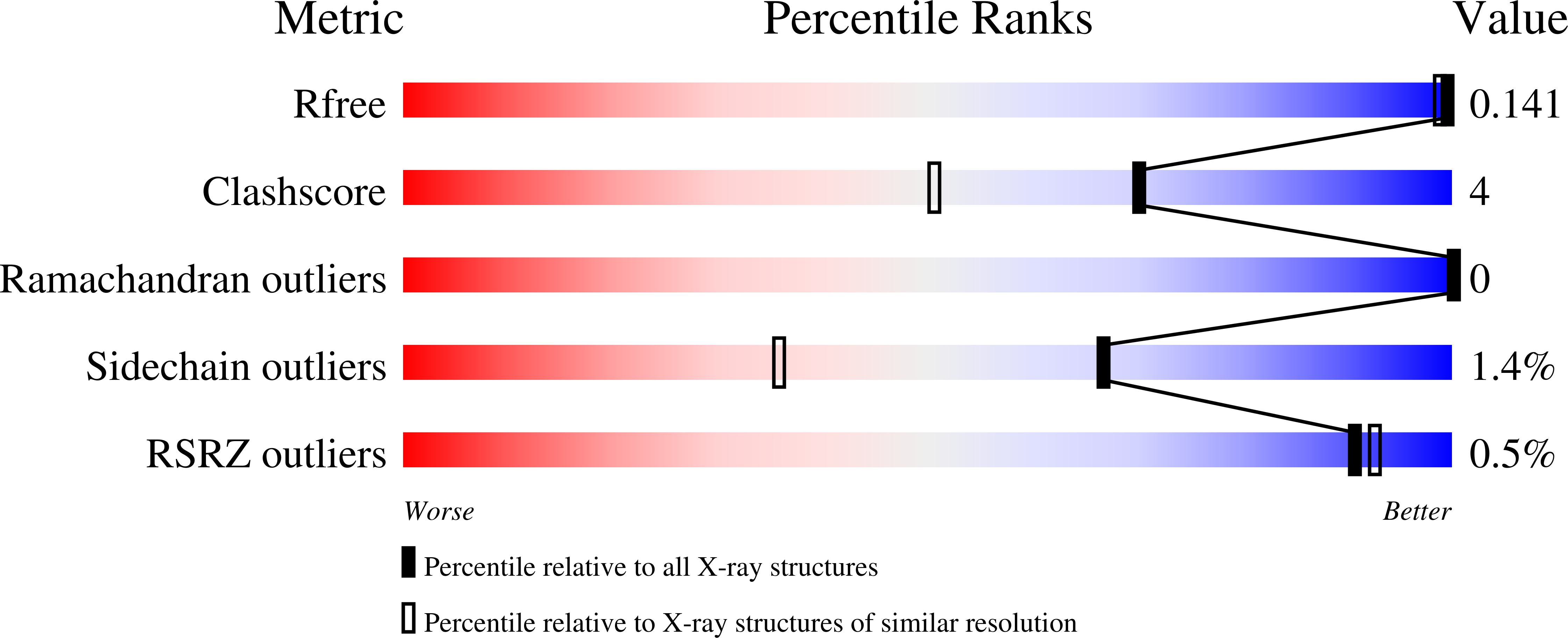
Resolution:
1.35 Å
R-Value Free:
0.14
R-Value Work:
0.12
Space Group:
P 21 21 21