Deposition Date
2018-08-13
Release Date
2018-11-21
Last Version Date
2024-03-06
Entry Detail
PDB ID:
5QJ0
Keywords:
Title:
CRYSTAL STRUCTURE OF THE HEPATITIS C VIRUS GENOTYPE 2A STRAIN JFH1 NS5B RNA-DEPENDENT RNA POLYMERASE IN COMPLEX WITH 6-[ethyl(methylsulfonyl)amino]-2-(4-fluorophenyl)-N-methyl-5-(3-{[1-(pyrimidin-2-yl)cyclopropyl]carbamoyl}phenyl)-1-benzofuran-3-carboxamide
Biological Source:
Source Organism(s):
Hepacivirus C (Taxon ID: 11103)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
2.08 Å
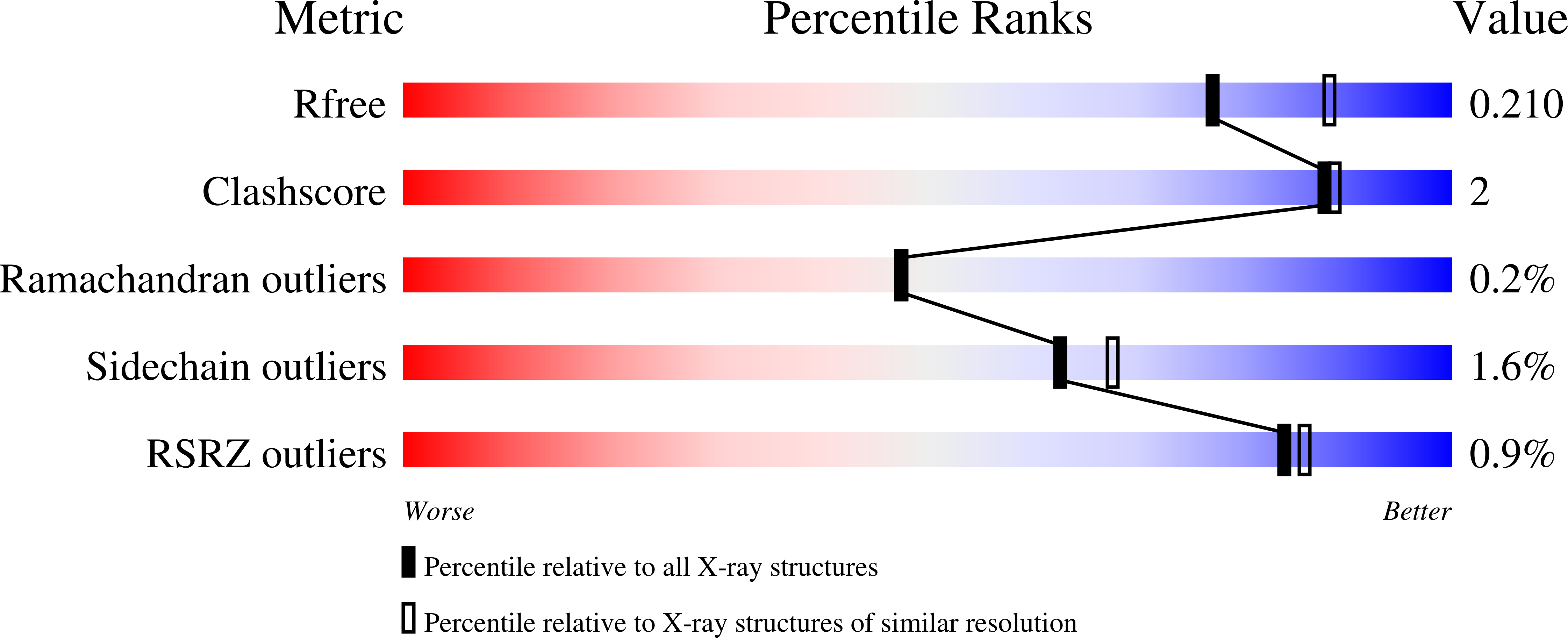
R-Value Free:
0.21
R-Value Work:
0.17
R-Value Observed:
0.17
Space Group:
P 21 21 21