Deposition Date
2017-08-04
Release Date
2017-11-29
Last Version Date
2024-01-17
Entry Detail
PDB ID:
5ONX
Keywords:
Title:
Resting state copper nitrite reductase determined by serial femtosecond rotation crystallography
Biological Source:
Source Organism(s):
Alcaligenes xylosoxydans xylosoxydans (Taxon ID: 85698)
Expression System(s):
Method Details:
Experimental Method:
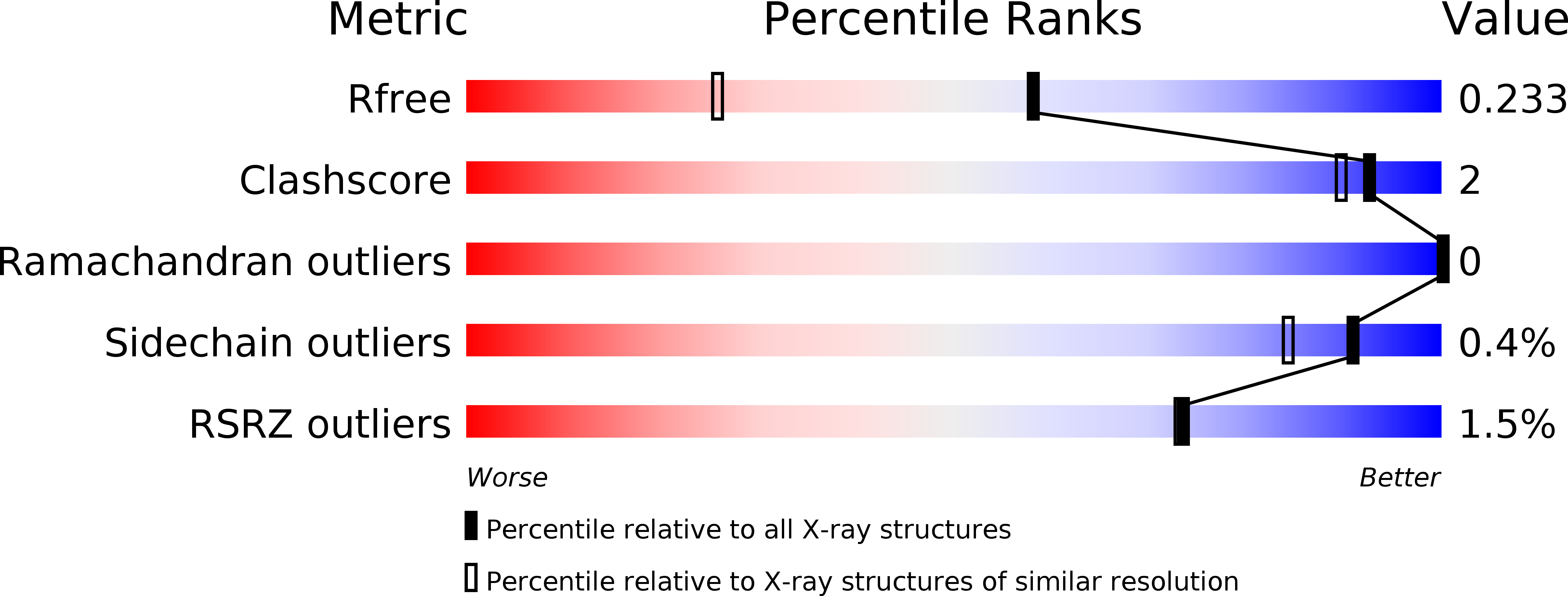
Resolution:
1.60 Å
R-Value Free:
0.22
R-Value Work:
0.18
R-Value Observed:
0.18
Space Group:
H 3