Deposition Date
2017-07-05
Release Date
2017-08-30
Last Version Date
2024-10-23
Entry Detail
PDB ID:
5ODH
Keywords:
Title:
Heterodisulfide reductase / [NiFe]-hydrogenase complex from Methanothermococcus thermolithotrophicus soaked with heterodisulfide for 3.5 minutes
Biological Source:
Source Organism(s):
Method Details:
Experimental Method:
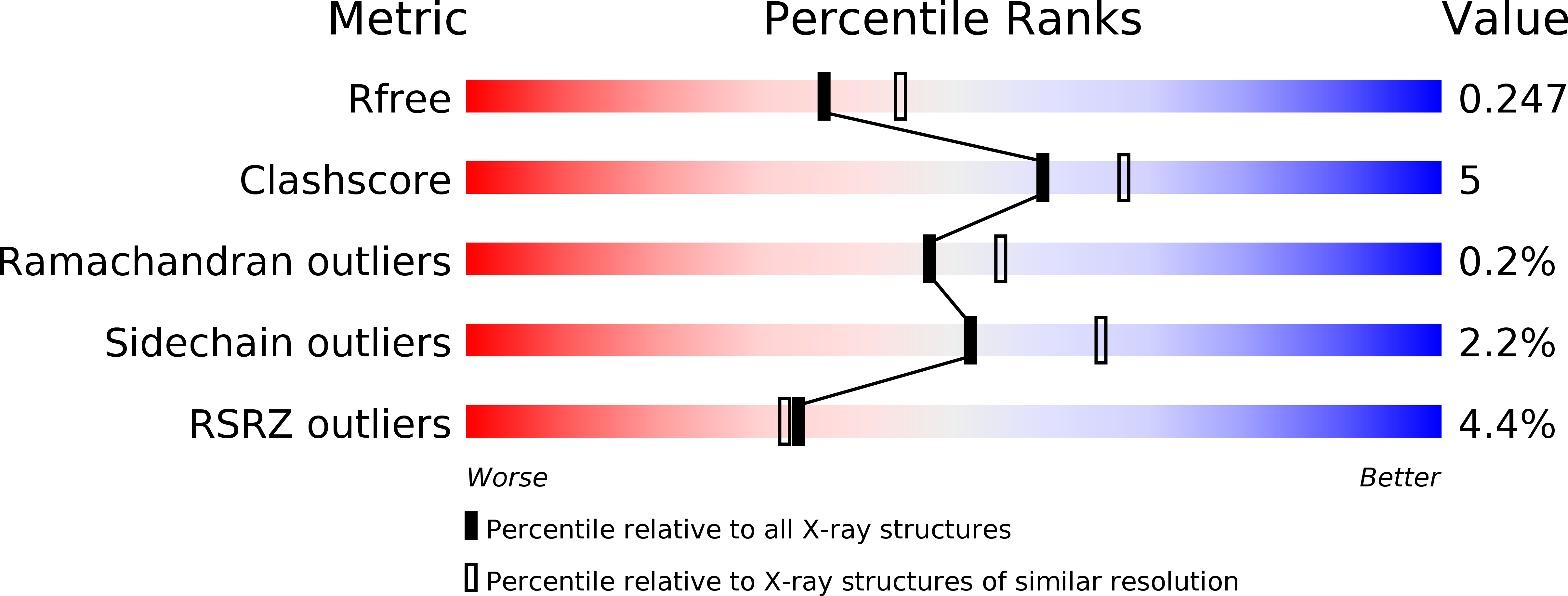
Resolution:
2.20 Å
R-Value Free:
0.24
R-Value Work:
0.21
R-Value Observed:
0.21
Space Group:
C 1 2 1