Deposition Date
2017-06-20
Release Date
2018-07-11
Last Version Date
2024-05-08
Entry Detail
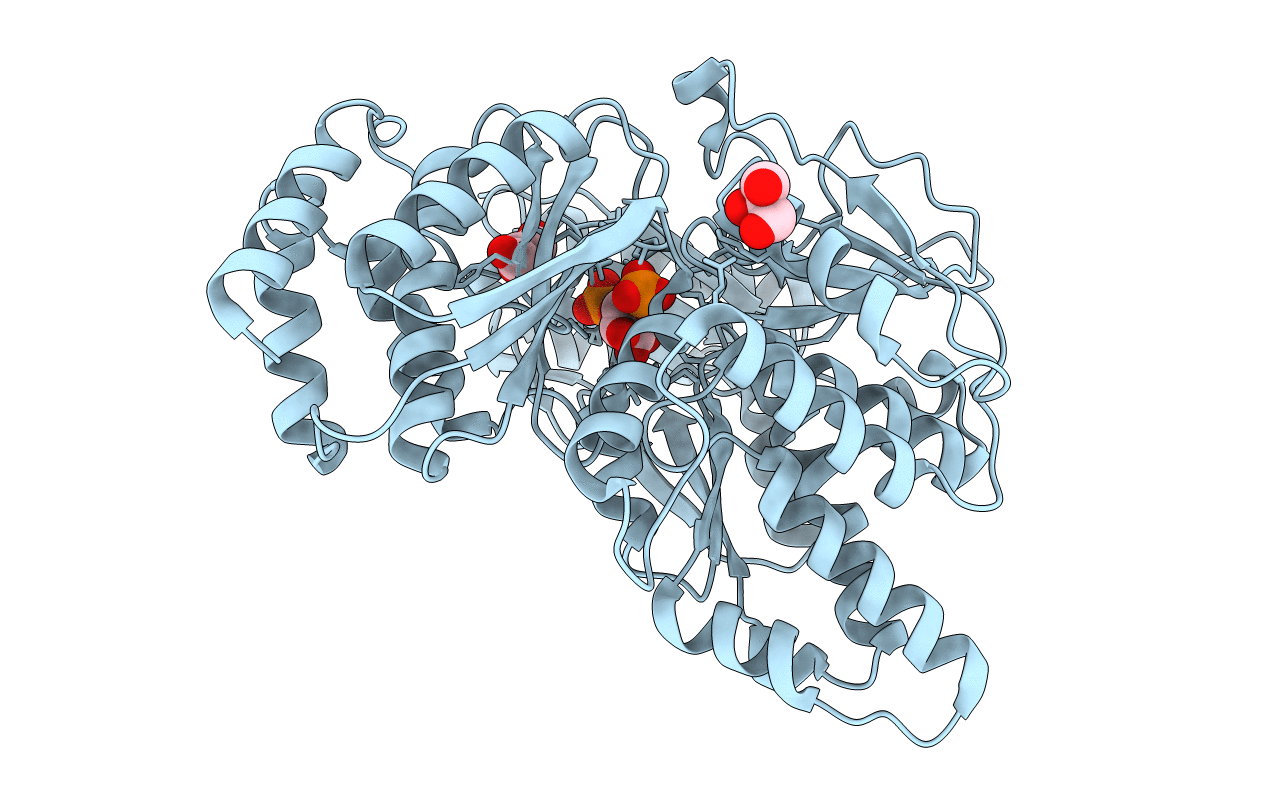
PDB ID:
5O9X
Keywords:
Title:
Crystal structure of Aspergillus fumigatus N-acetylphosphoglucosamine mutate S69A in complex with glucose1,6bisphosphate
Biological Source:
Source Organism:
Aspergillus lentulus (Taxon ID: 293939)
Host Organism:
Method Details:
Experimental Method:
Resolution:
1.90 Å
R-Value Free:
0.20
R-Value Work:
0.16
R-Value Observed:
0.16
Space Group:
P 21 21 21


