Deposition Date
2017-02-16
Release Date
2017-04-12
Last Version Date
2024-10-23
Entry Detail
PDB ID:
5N6U
Keywords:
Title:
Crystal structure of Beta-D-Mannosidase from Dictyoglomus thermophilum.
Biological Source:
Source Organism:
Dictyoglomus thermophilum H-6-12 (Taxon ID: 309799)
Host Organism:
Method Details:
Experimental Method:
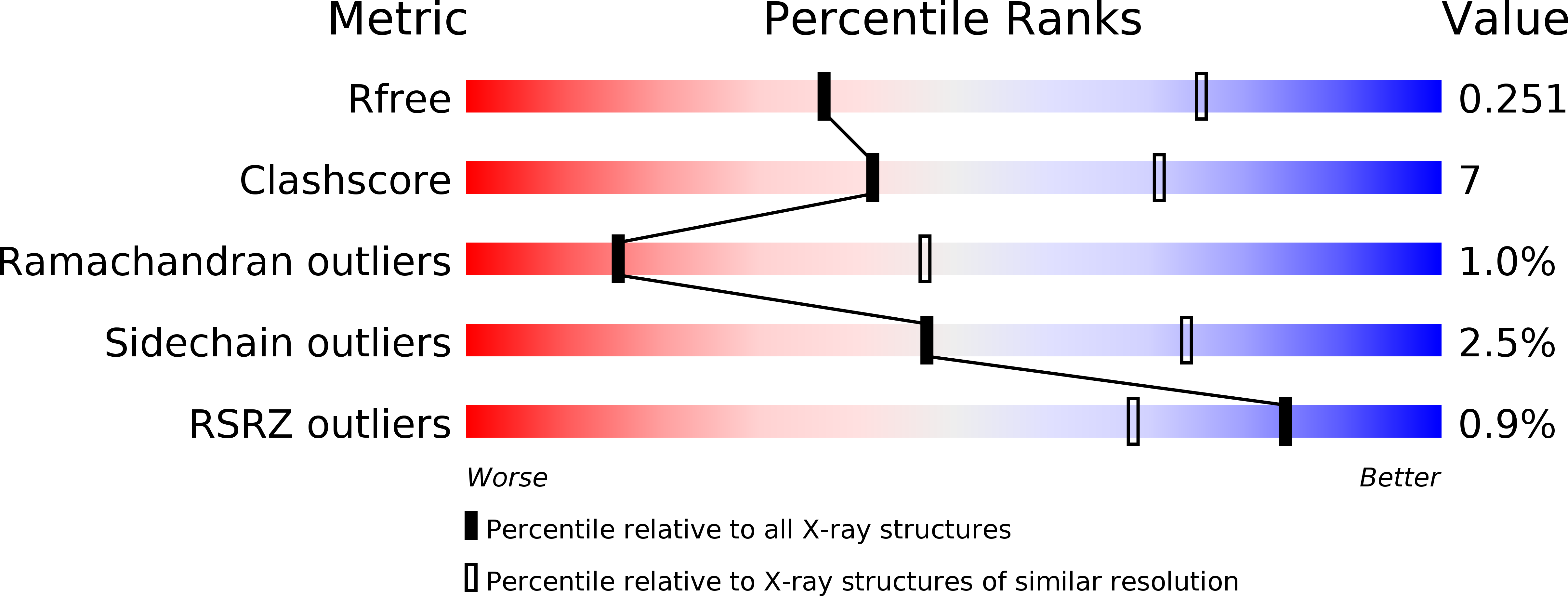
Resolution:
3.08 Å
R-Value Free:
0.25
R-Value Work:
0.20
R-Value Observed:
0.20
Space Group:
P 1 21 1