Deposition Date
2017-02-10
Release Date
2017-11-01
Last Version Date
2024-05-08
Entry Detail
PDB ID:
5N4C
Keywords:
Title:
Prolyl oligopeptidase B from Galerina marginata bound to 35mer hydrolysis and macrocyclization substrate - S577A mutant
Biological Source:
Source Organism:
Galerina marginata (Taxon ID: 109633)
Host Organism:
Method Details:
Experimental Method:
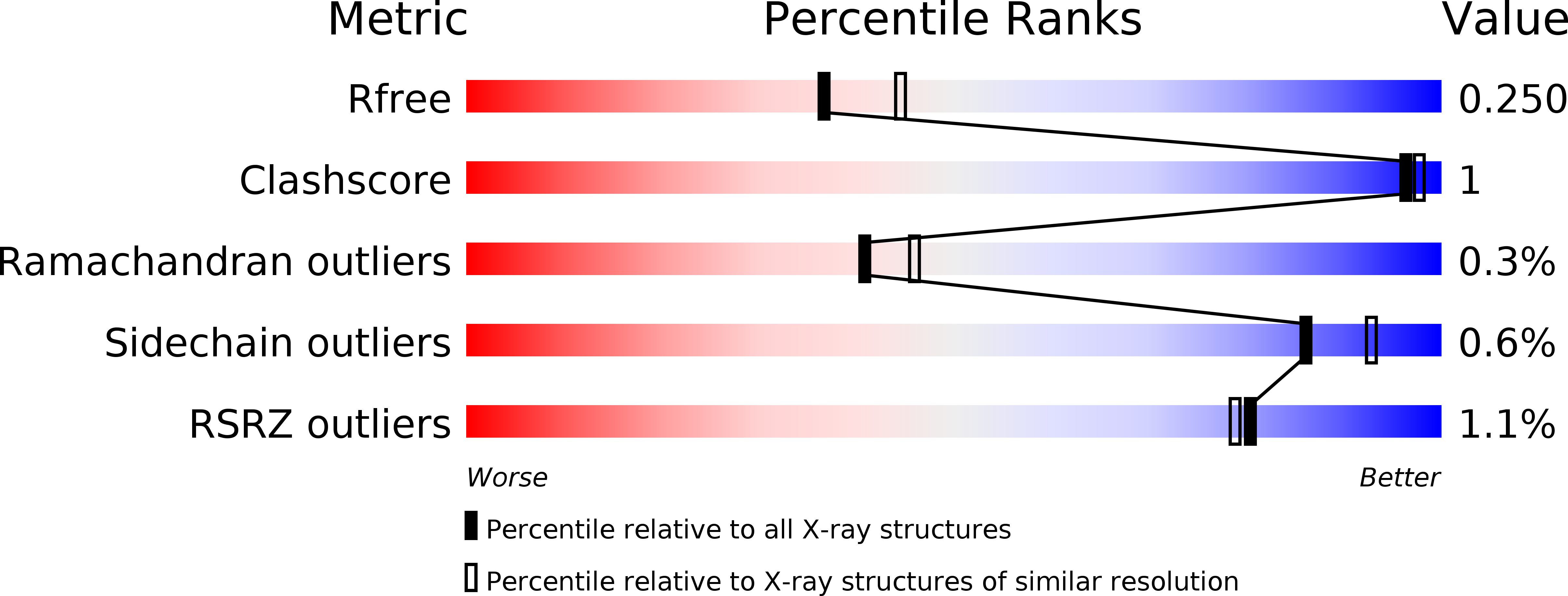
Resolution:
2.19 Å
R-Value Free:
0.24
R-Value Work:
0.21
R-Value Observed:
0.21
Space Group:
P 1 21 1