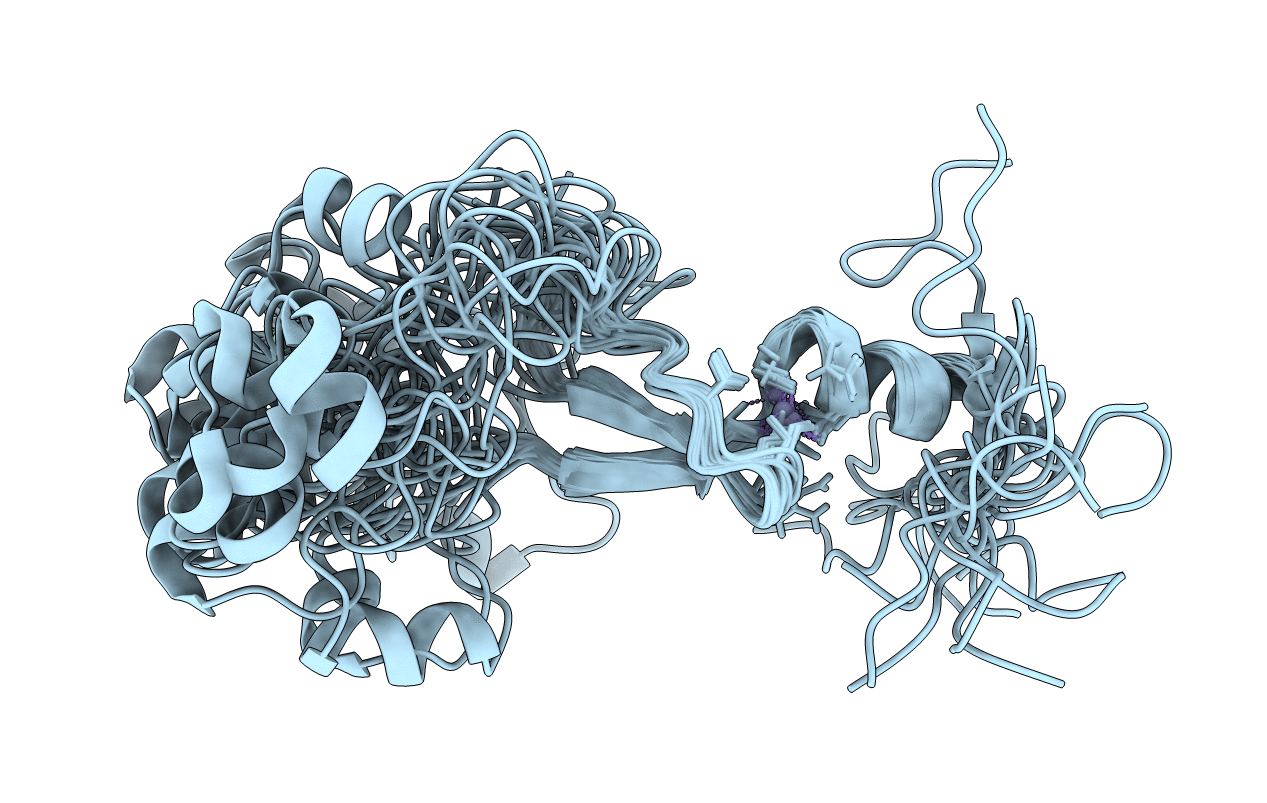
Deposition Date
2017-01-05
Release Date
2017-06-21
Last Version Date
2024-06-19
Entry Detail
PDB ID:
5MSL
Keywords:
Title:
Solution structure of the B. subtilis anti-sigma-F factor, FIN
Biological Source:
Source Organism(s):
Bacillus subtilis (Taxon ID: 1423)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
100
Conformers Submitted:
20
Selection Criteria:
structures with the lowest energy