Deposition Date
2016-11-14
Release Date
2017-01-11
Last Version Date
2025-10-01
Entry Detail
PDB ID:
5ME0
Keywords:
Title:
Structure of the 30S Pre-Initiation Complex 1 (30S IC-1) Stalled by GE81112
Biological Source:
Source Organism(s):
Thermus thermophilus (strain HB8 / ATCC 27634 / DSM 579) (Taxon ID: 300852)
Escherichia coli K-12 (Taxon ID: 83333)
Geobacillus stearothermophilus (Taxon ID: 1422)
Escherichia coli K-12 (Taxon ID: 83333)
Geobacillus stearothermophilus (Taxon ID: 1422)
Expression System(s):
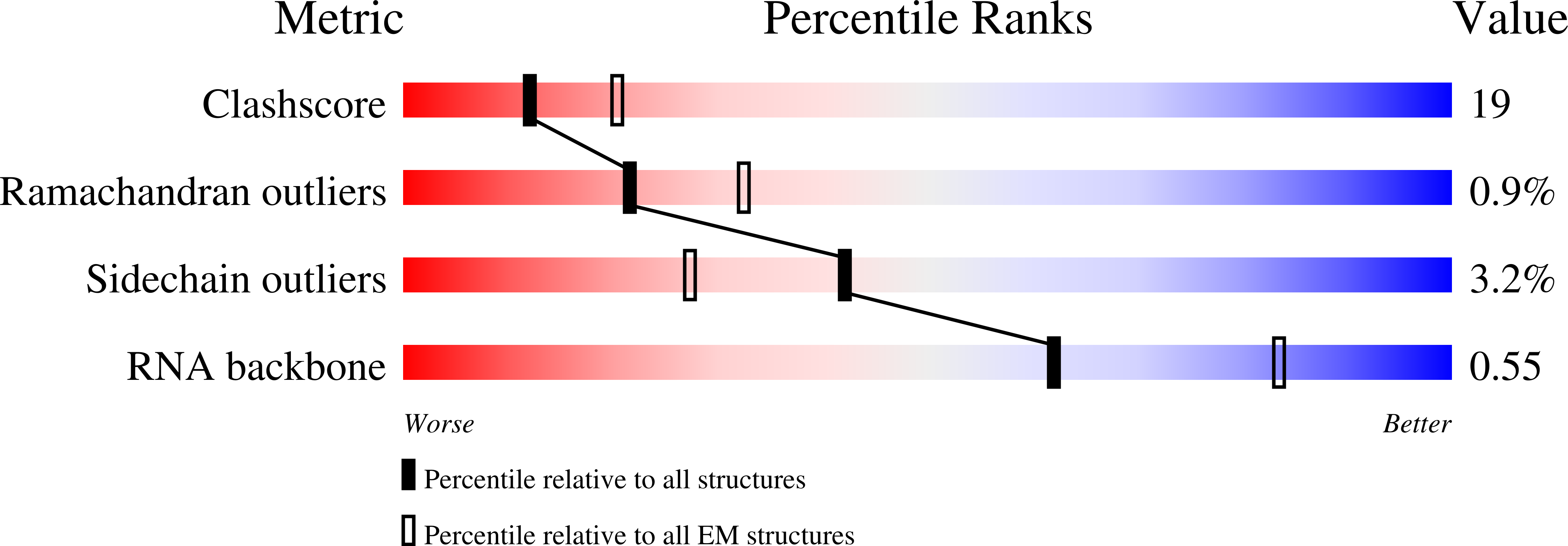
Method Details:
Experimental Method:
Resolution:
13.50 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE