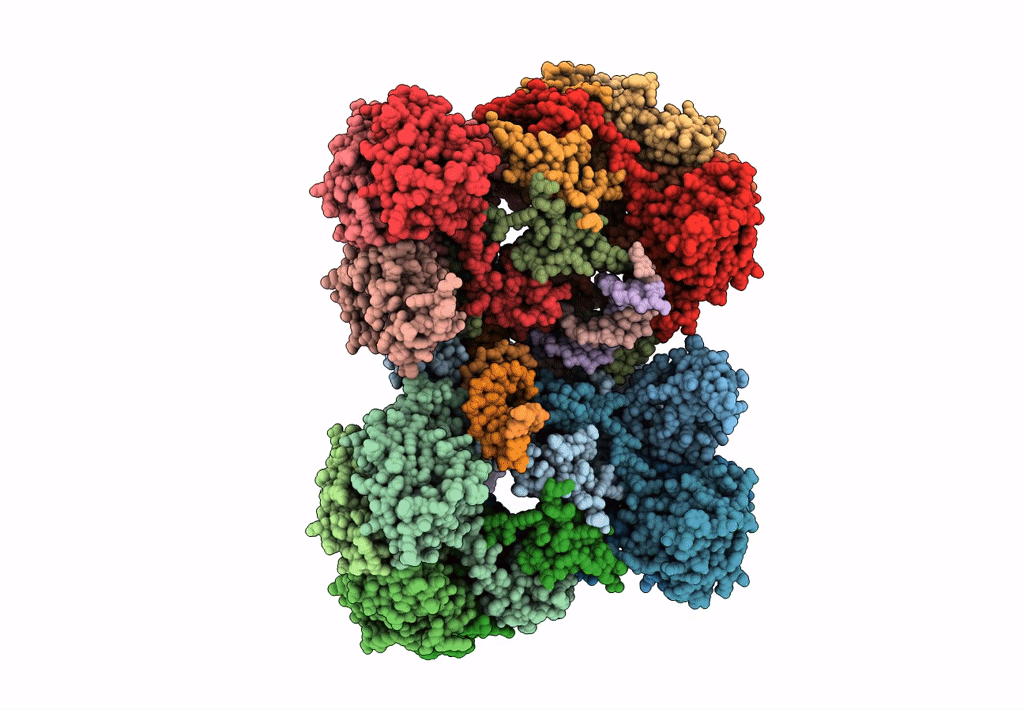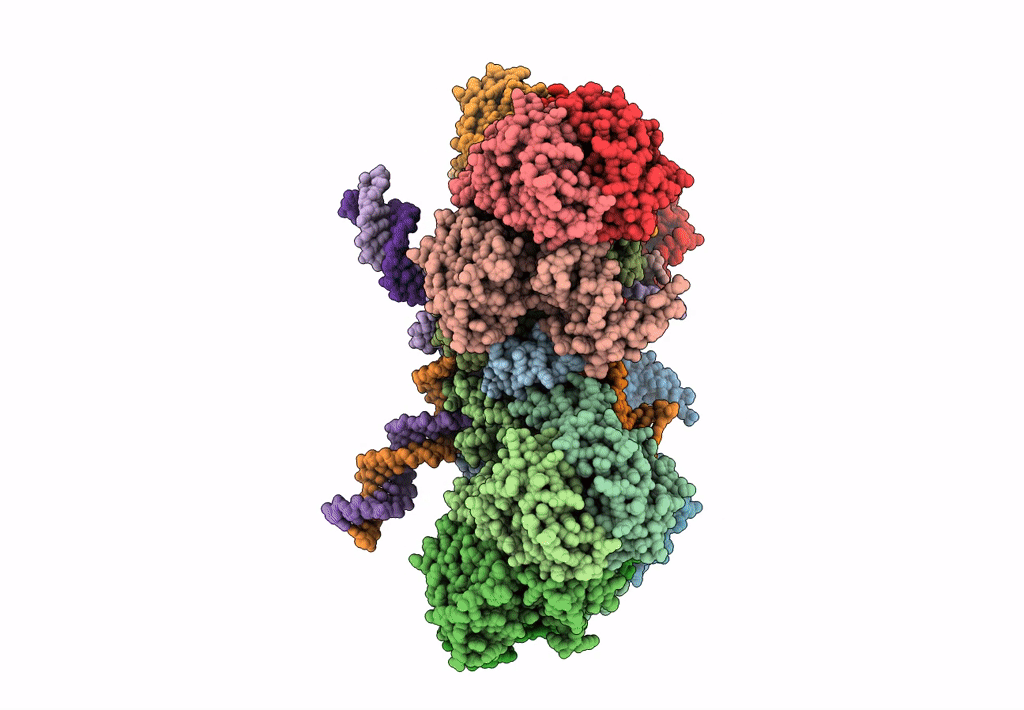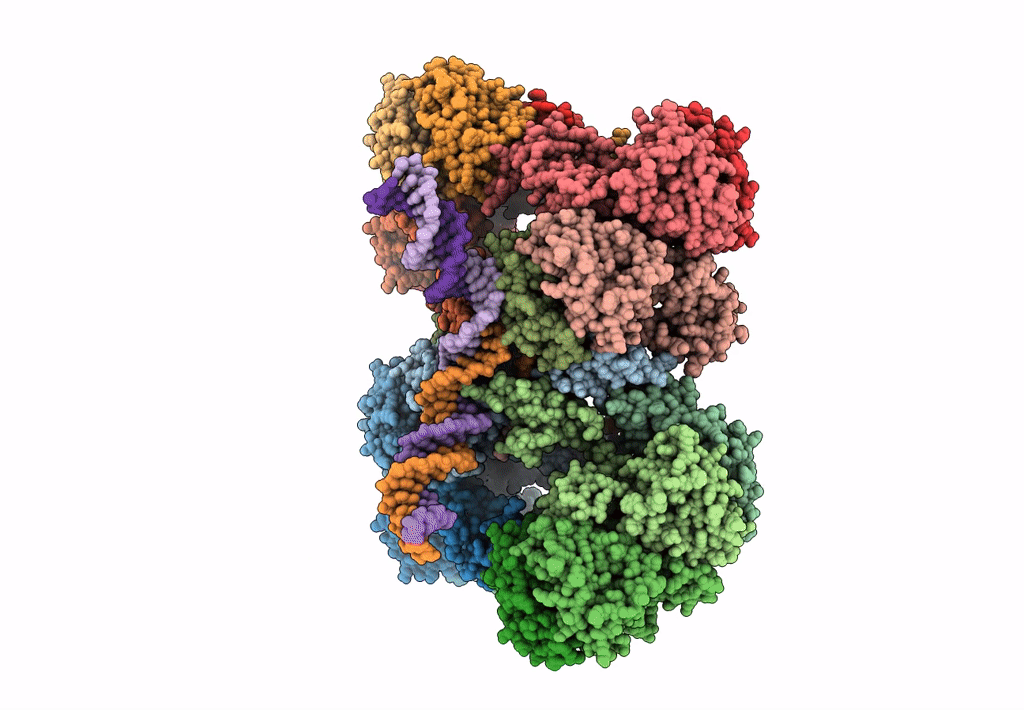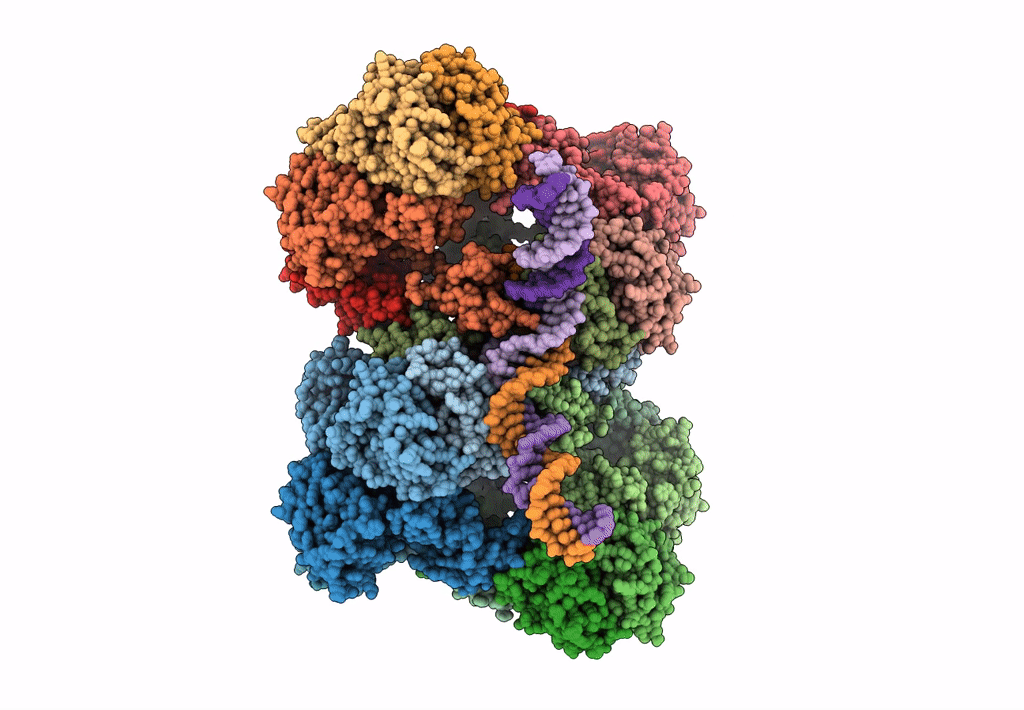
Deposition Date
2016-10-05
Release Date
2017-01-18
Last Version Date
2024-05-15
Entry Detail
PDB ID:
5M0R
Keywords:
Title:
Cryo-EM reconstruction of the maedi-visna virus (MVV) strand transfer complex
Biological Source:
Source Organism(s):
Maedi visna virus (strain KV1772) (Taxon ID: 36374)
Visna lentivirus (strain 1514) (Taxon ID: 11742)
synthetic construct (Taxon ID: 32630)
Visna lentivirus (strain 1514) (Taxon ID: 11742)
synthetic construct (Taxon ID: 32630)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
8.20 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE


