Deposition Date
2016-08-25
Release Date
2016-11-09
Last Version Date
2024-05-01
Entry Detail
PDB ID:
5LSE
Keywords:
Title:
PHOTOSYNTHETIC REACTION CENTER MUTANT WITH Glu L212 replaced with Ala (CHAIN L, EL212W), Asp L213 replaced with ALA (Chain L, DL213A) AND LEU M215 REPLACED WITH ALA (CHAIN M, LM215A)
Biological Source:
Source Organism(s):
Rhodobacter sphaeroides (Taxon ID: 1063)
Expression System(s):
Method Details:
Experimental Method:
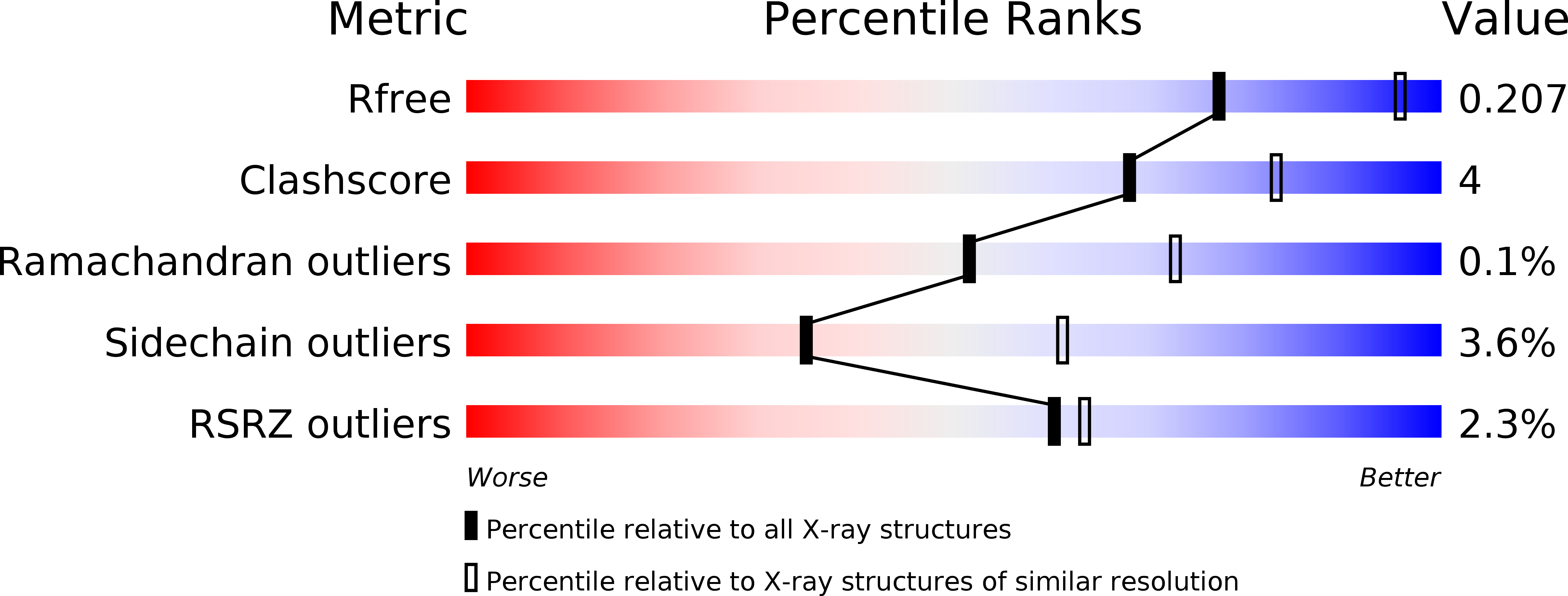
Resolution:
2.50 Å
R-Value Free:
0.20
R-Value Work:
0.16
R-Value Observed:
0.16
Space Group:
P 31 2 1