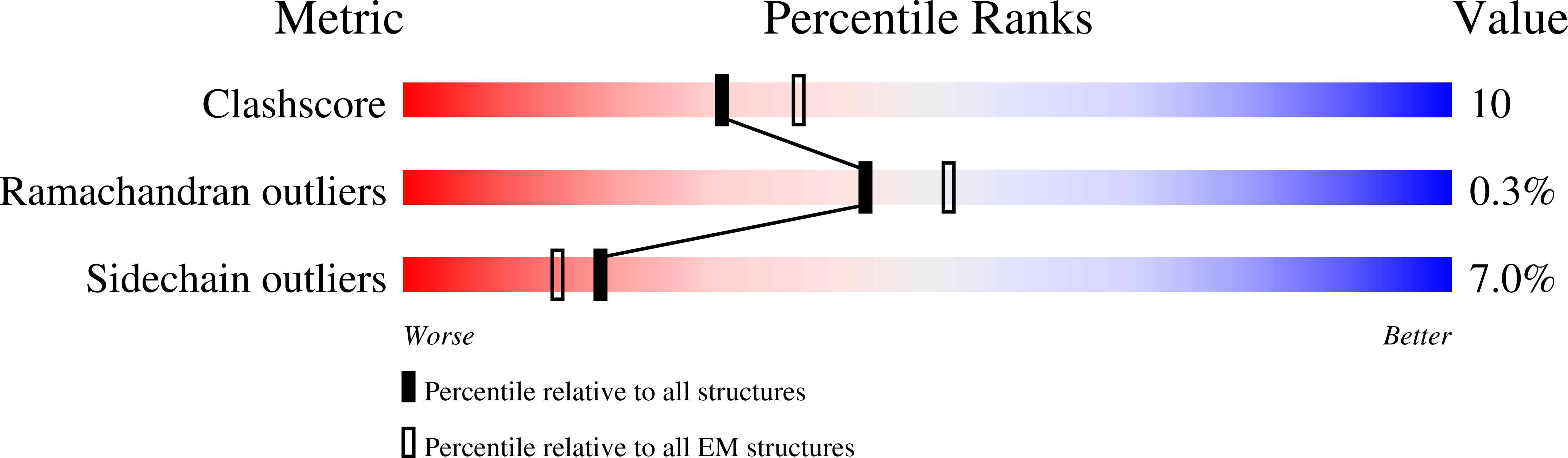
Deposition Date
2016-07-21
Release Date
2017-01-18
Last Version Date
2024-05-15
Entry Detail
PDB ID:
5LK8
Keywords:
Title:
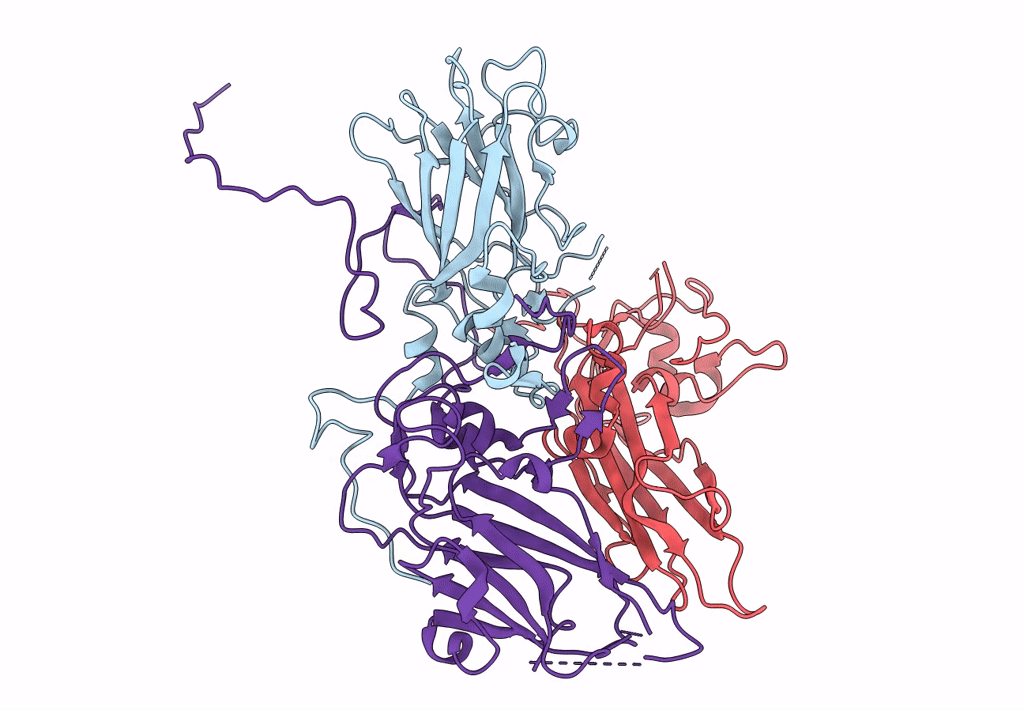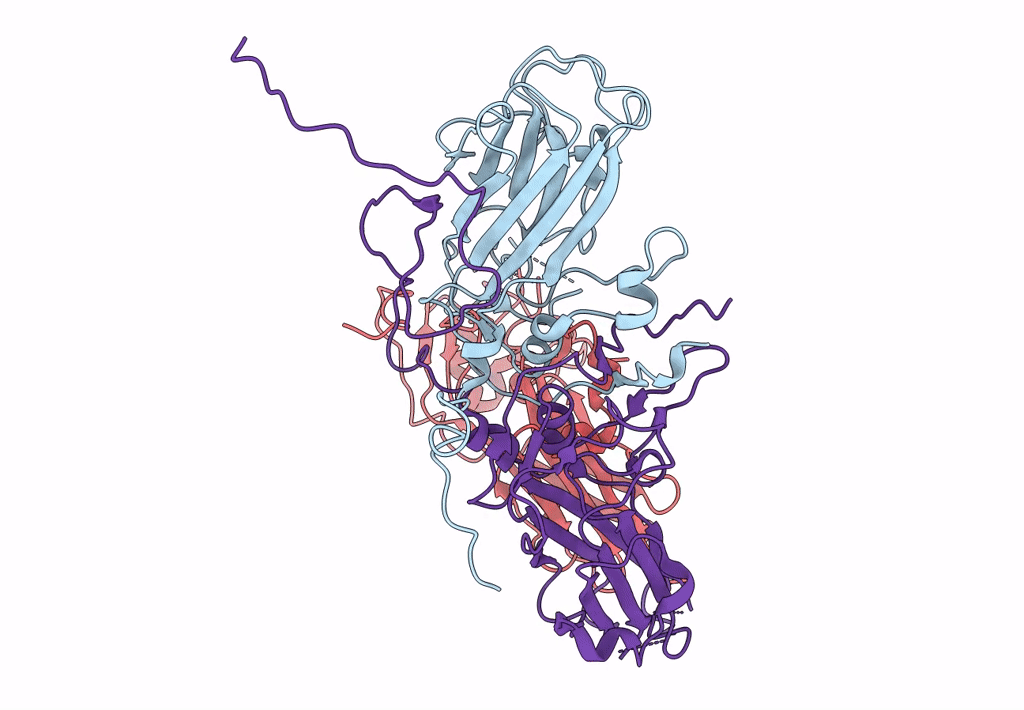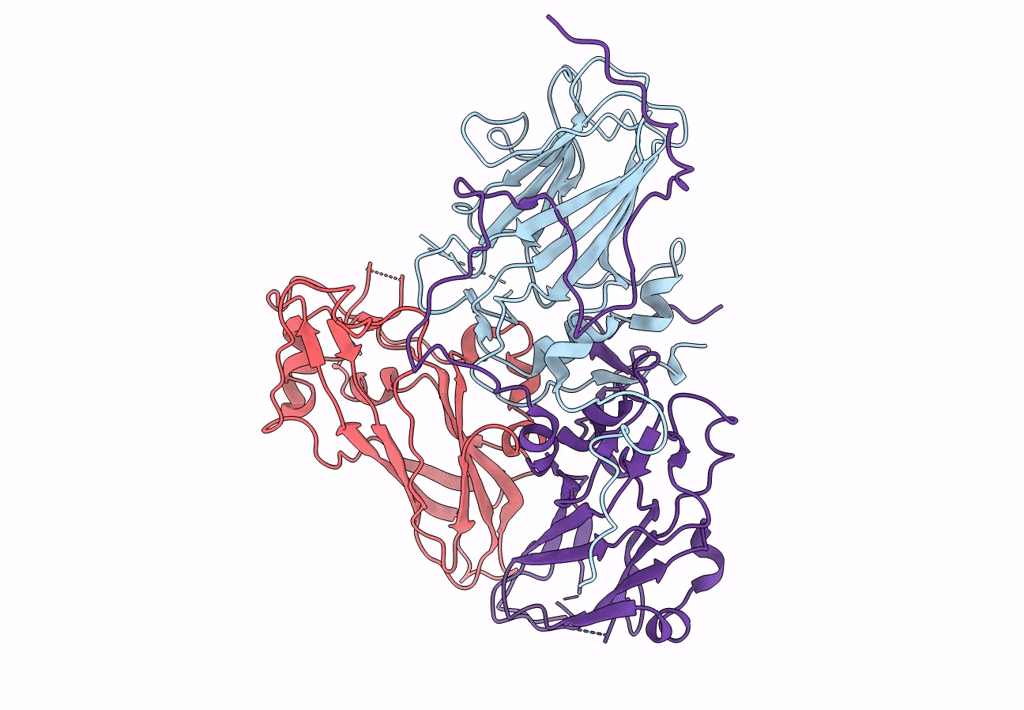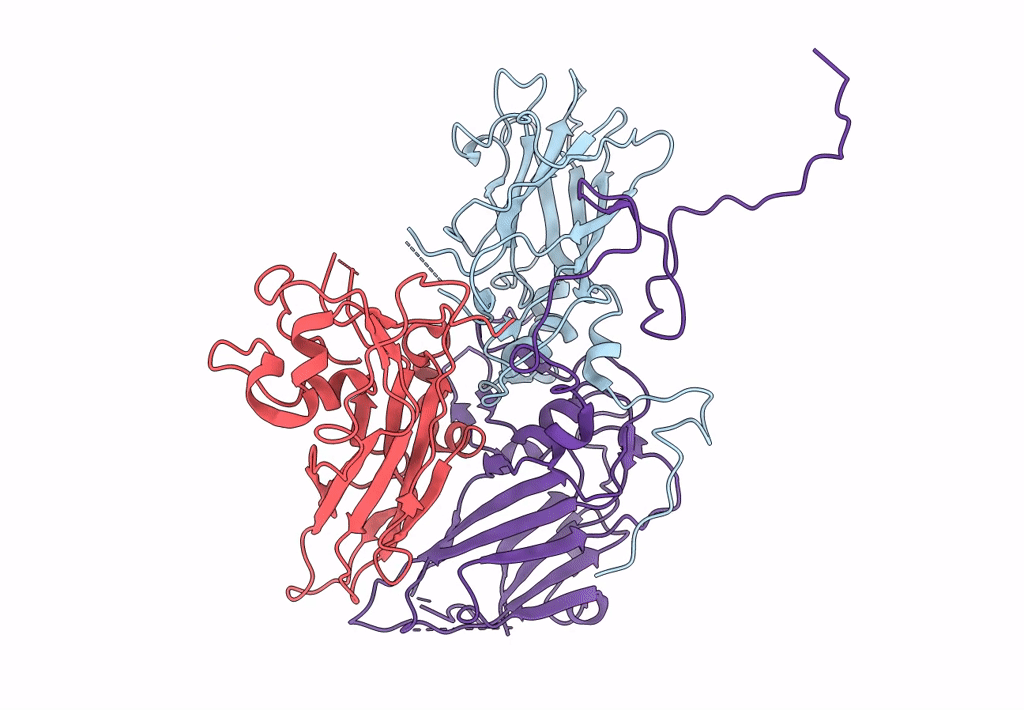
single particle reconstruction of slow bee paralysis virus empty particle
Biological Source:
Source Organism(s):
Slow bee paralysis virus (Taxon ID: 458132)
Method Details:
Experimental Method:
Resolution:
3.42 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE