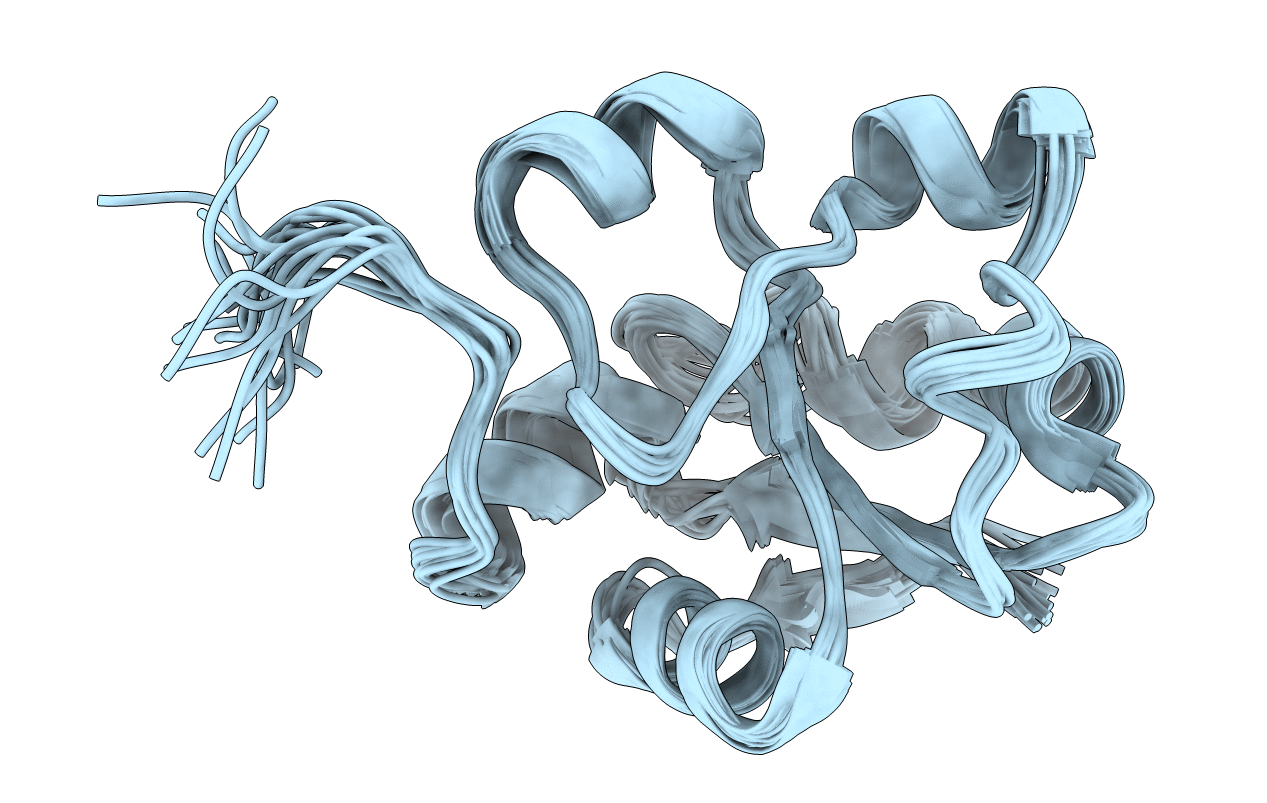
Deposition Date
2016-06-14
Release Date
2016-08-17
Last Version Date
2024-06-19
Entry Detail
PDB ID:
5LAO
Keywords:
Title:
S-nitrosylated 3D NMR structure of the cytoplasmic rhodanese domain of the inner membrane protein YgaP from Escherichia coli
Biological Source:
Source Organism(s):
Escherichia coli (strain K12) (Taxon ID: 83333)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
100
Conformers Submitted:
20
Selection Criteria:
structures with the least restraint violations


