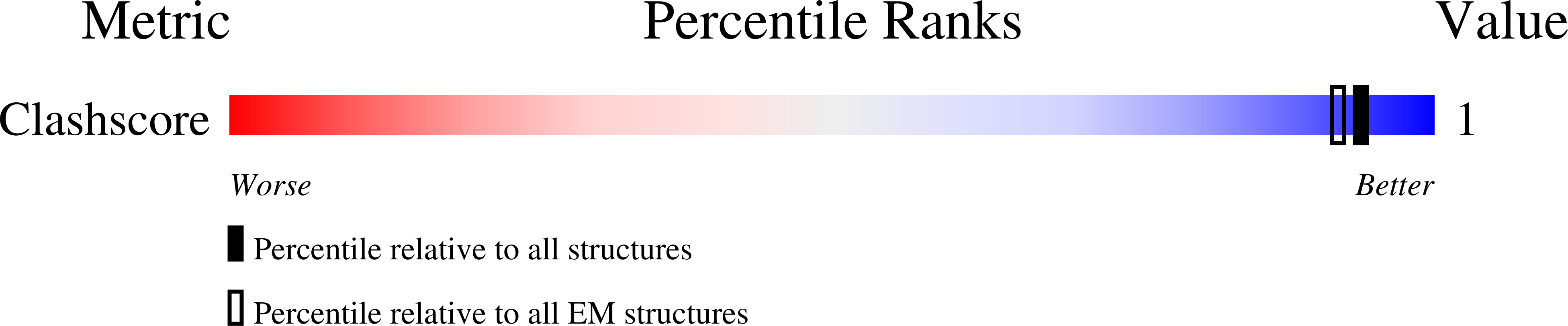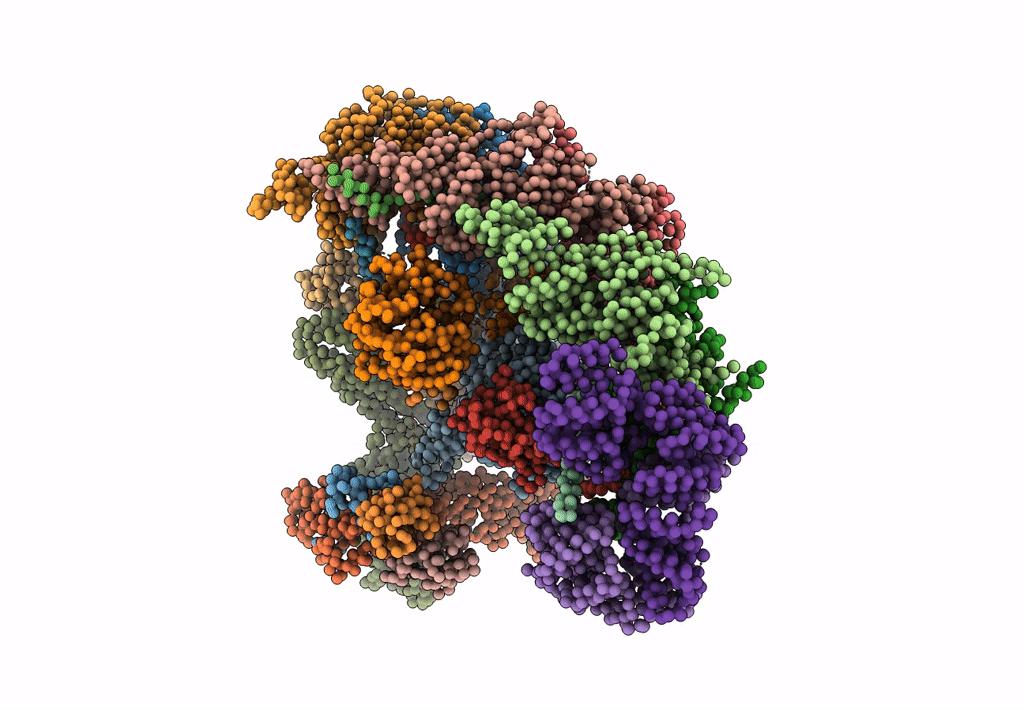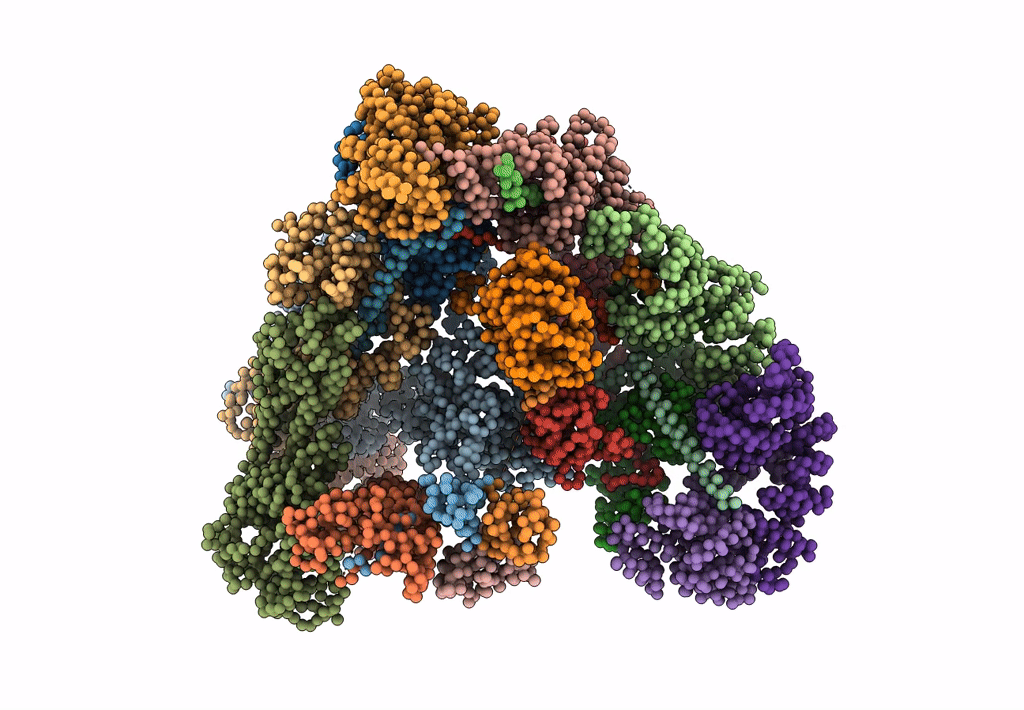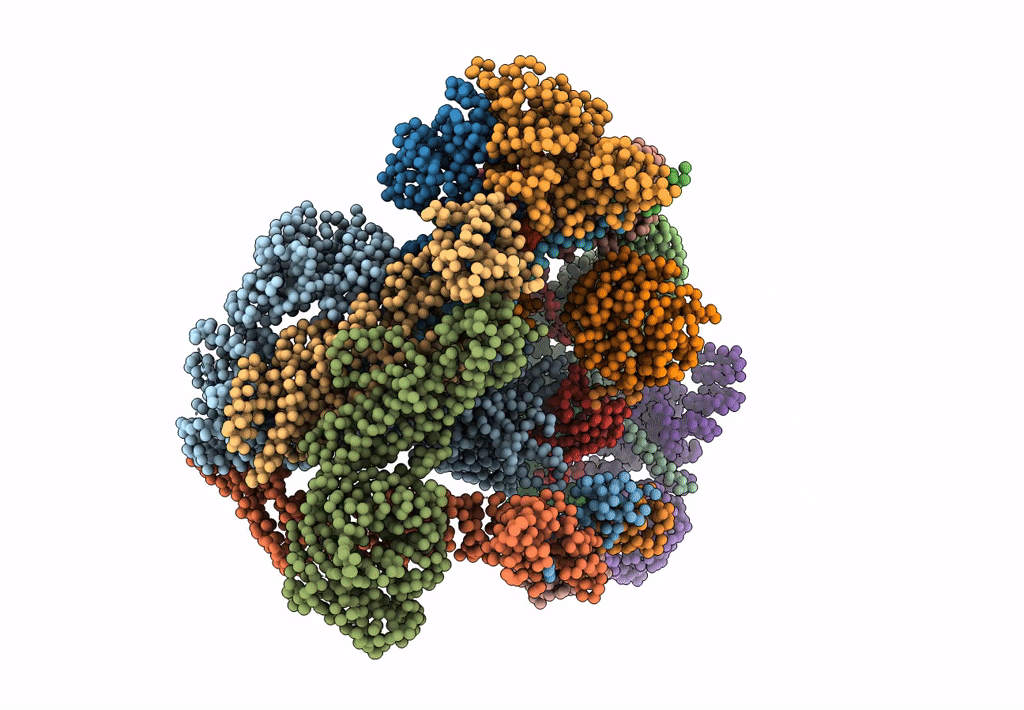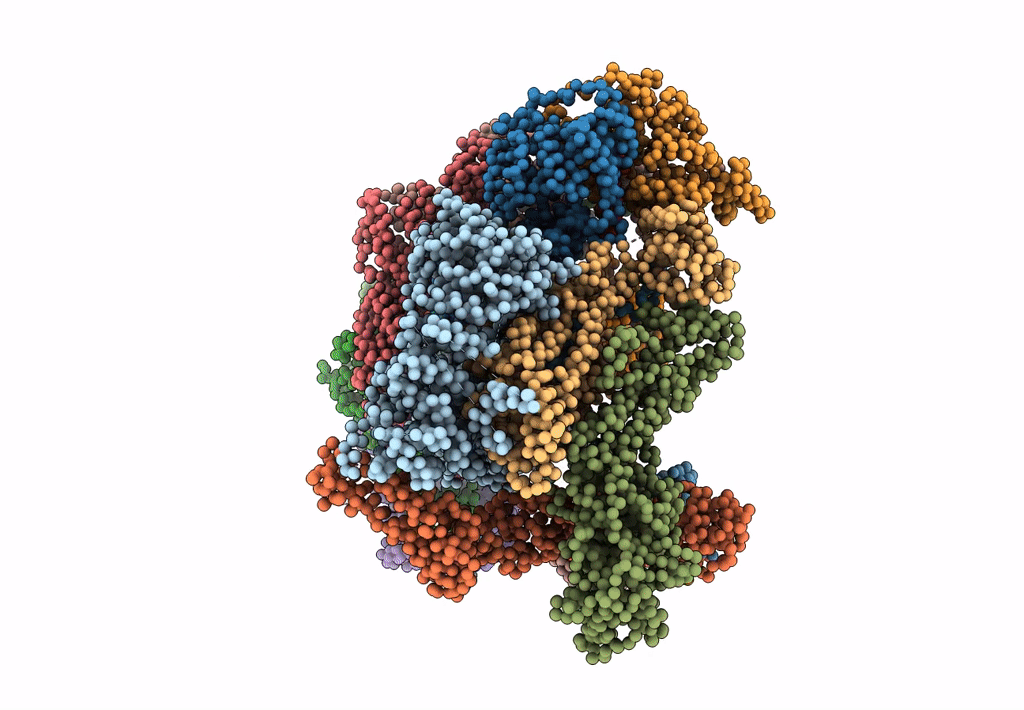
Deposition Date
2016-06-11
Release Date
2016-10-26
Last Version Date
2024-05-08
Entry Detail
PDB ID:
5L9T
Keywords:
Title:
Model of human Anaphase-promoting complex/Cyclosome (APC/C-CDH1) with E2 UBE2S poised for polyubiquitination where UBE2S, APC2, and APC11 are modeled into low resolution density
Biological Source:
Source Organism(s):
Homo sapiens (Taxon ID: 9606)
Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Taxon ID: 559292)
Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Taxon ID: 559292)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
6.40 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE