Deposition Date
2016-07-09
Release Date
2017-07-19
Last Version Date
2023-10-04
Entry Detail
PDB ID:
5KSS
Keywords:
Title:
Stationary phase survival protein E (SurE) from Xylella fastidiosa - XFSurE-Ds (Dimer Smaller)
Biological Source:
Source Organism(s):
Xylella fastidiosa (strain 9a5c) (Taxon ID: 160492)
Expression System(s):
Method Details:
Experimental Method:
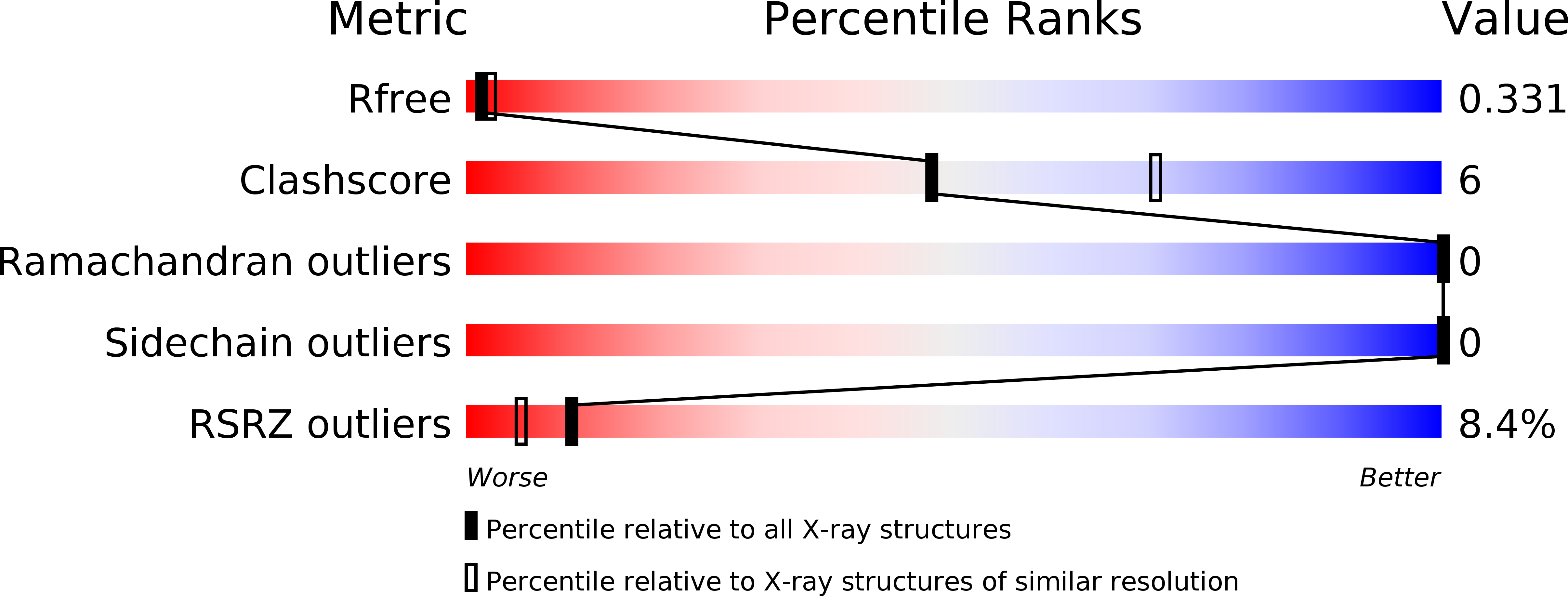
Resolution:
2.82 Å
R-Value Free:
0.32
R-Value Work:
0.27
R-Value Observed:
0.27
Space Group:
C 1 2 1