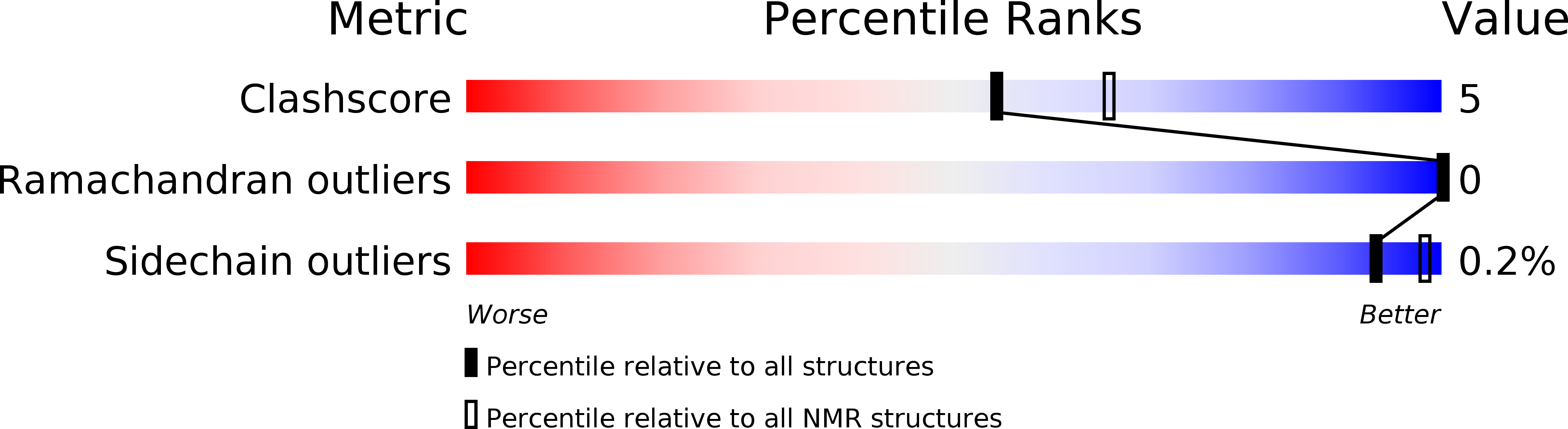Deposition Date
2016-07-06
Release Date
2017-07-12
Last Version Date
2024-05-15
Entry Detail
PDB ID:
5KQJ
Keywords:
Title:
Solution Structure of Antibiotic-Resistance Factor ANT(2'')-Ia Reveals Substrate-Regulated Conformation Dynamics
Biological Source:
Source Organism(s):
Klebsiella pneumoniae (Taxon ID: 573)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
15
Conformers Submitted:
15
Selection Criteria:
back calculated data agree with experimental NOESY spectrum