Deposition Date
2016-05-28
Release Date
2016-06-15
Last Version Date
2023-09-27
Entry Detail
PDB ID:
5K8B
Keywords:
Title:
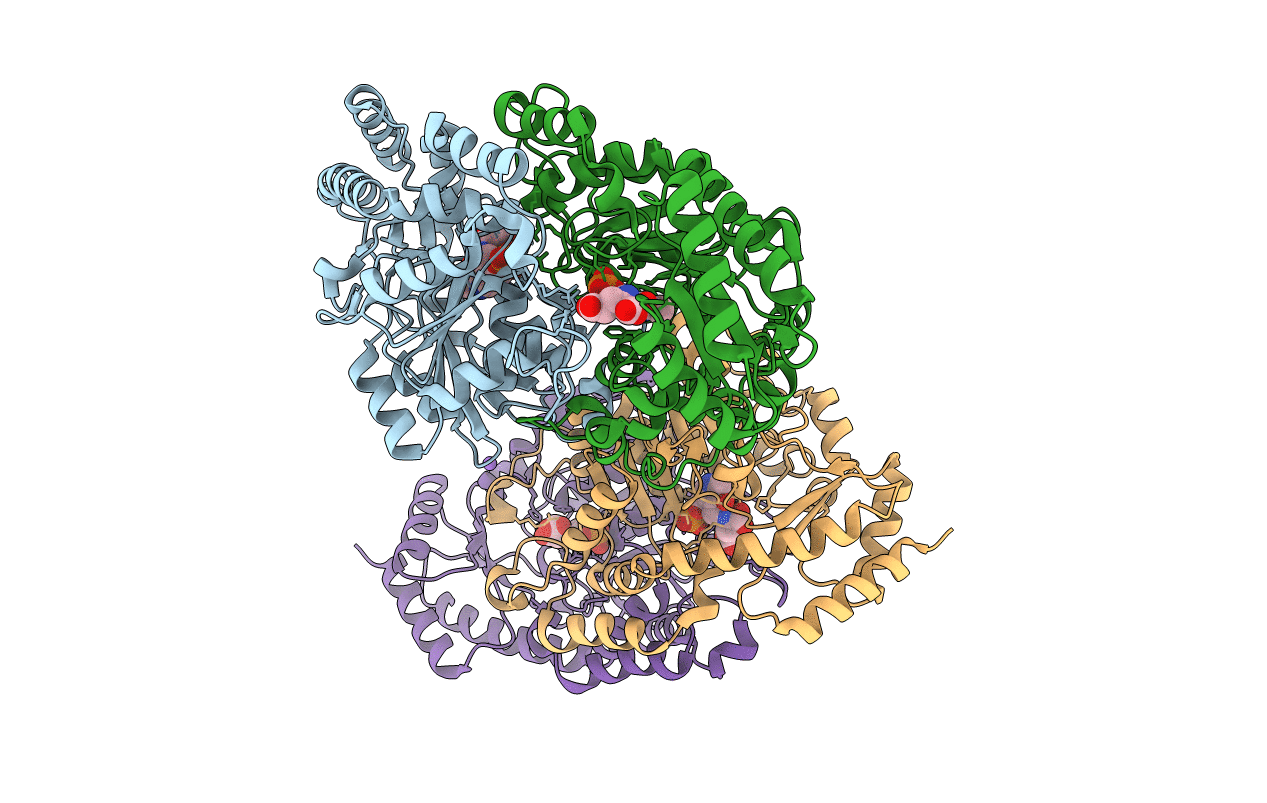
X-ray structure of KdnA, 8-amino-3,8-dideoxy-alpha-D-manno-octulosonate transaminase, from Shewanella oneidensis in the presence of the external aldimine with PLP and glutamate
Biological Source:
Source Organism(s):
Shewanella oneidensis (strain MR-1) (Taxon ID: 211586)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
2.15 Å
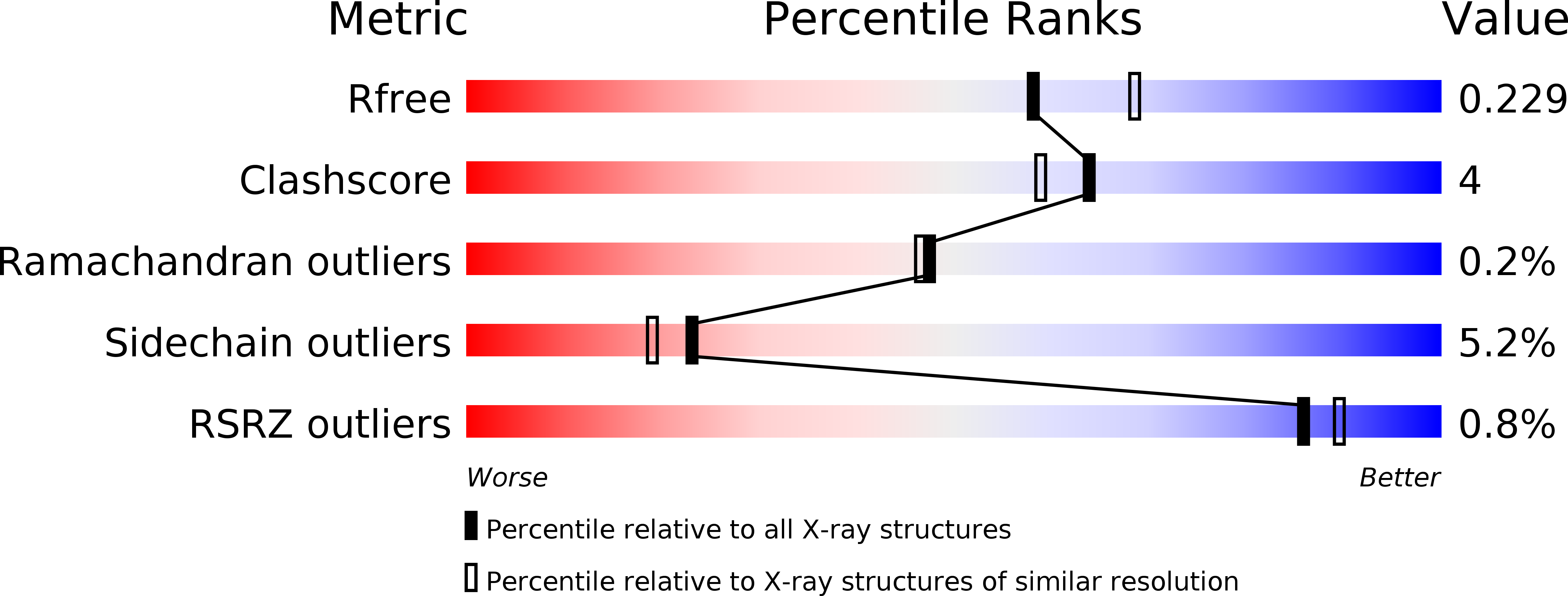
R-Value Free:
0.22
R-Value Work:
0.17
R-Value Observed:
0.17
Space Group:
C 1 2 1