Deposition Date
2016-05-10
Release Date
2016-10-05
Last Version Date
2025-04-02
Entry Detail
PDB ID:
5JUP
Keywords:
Title:
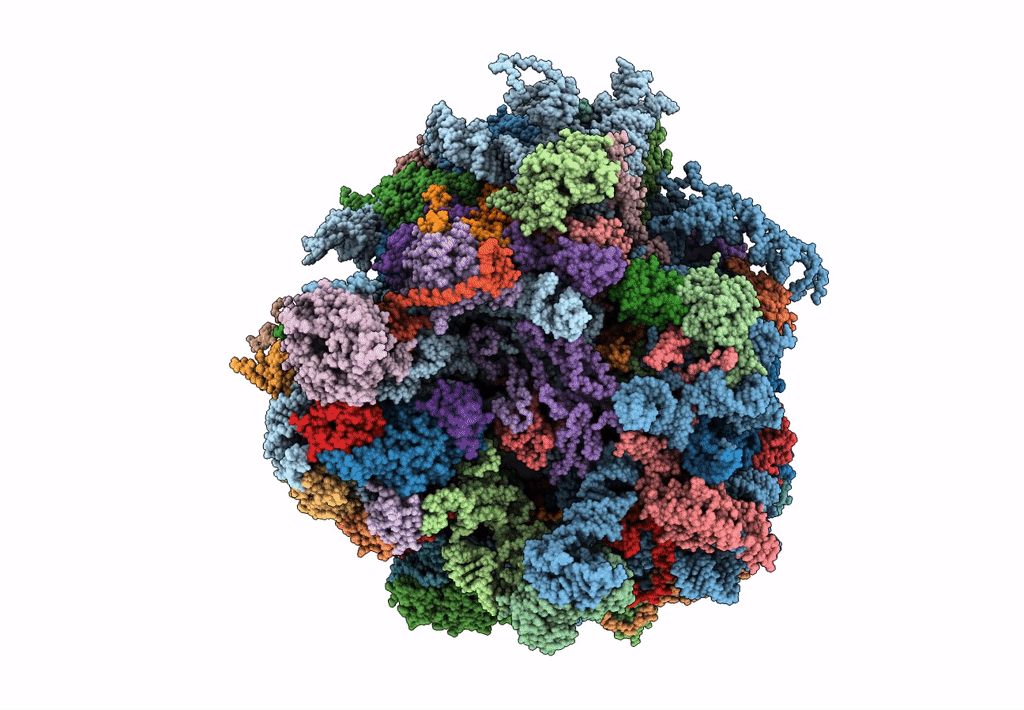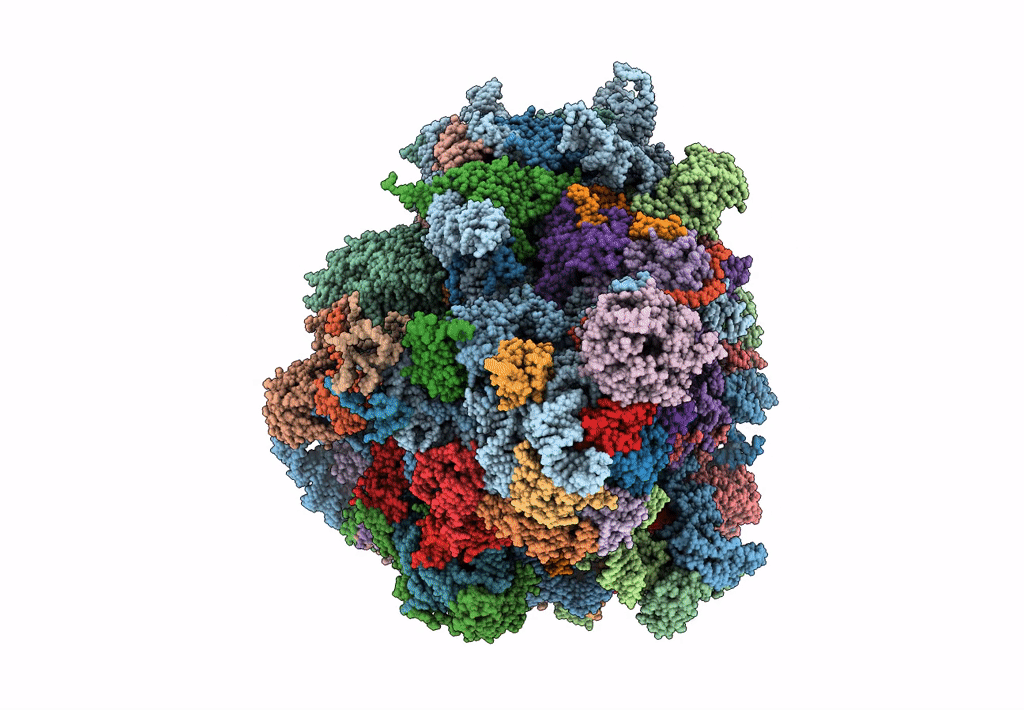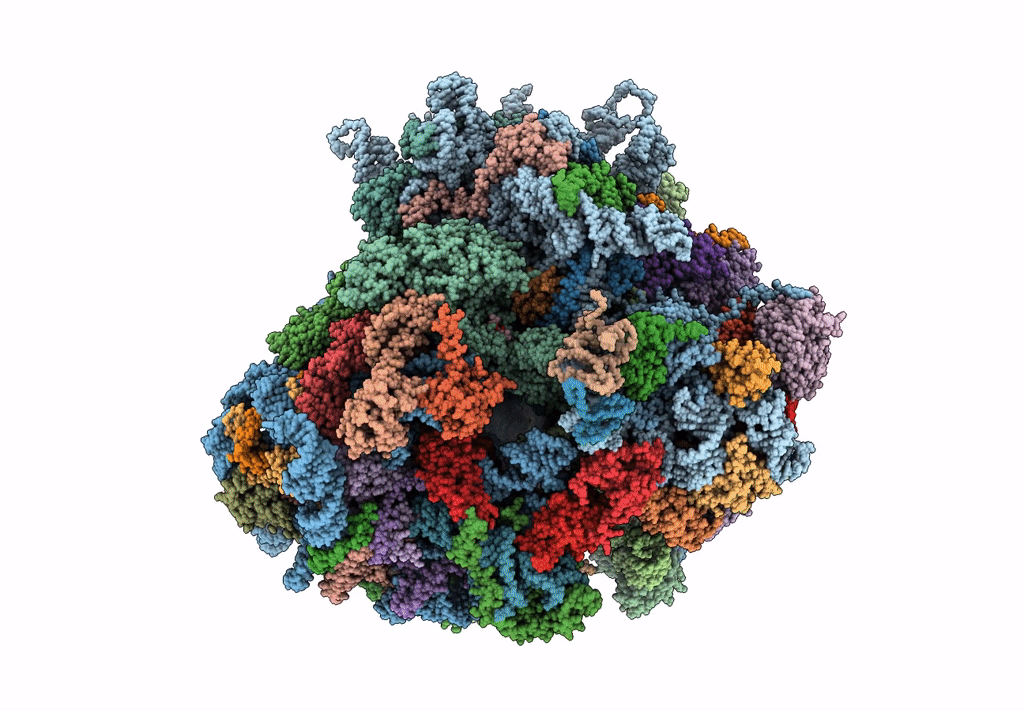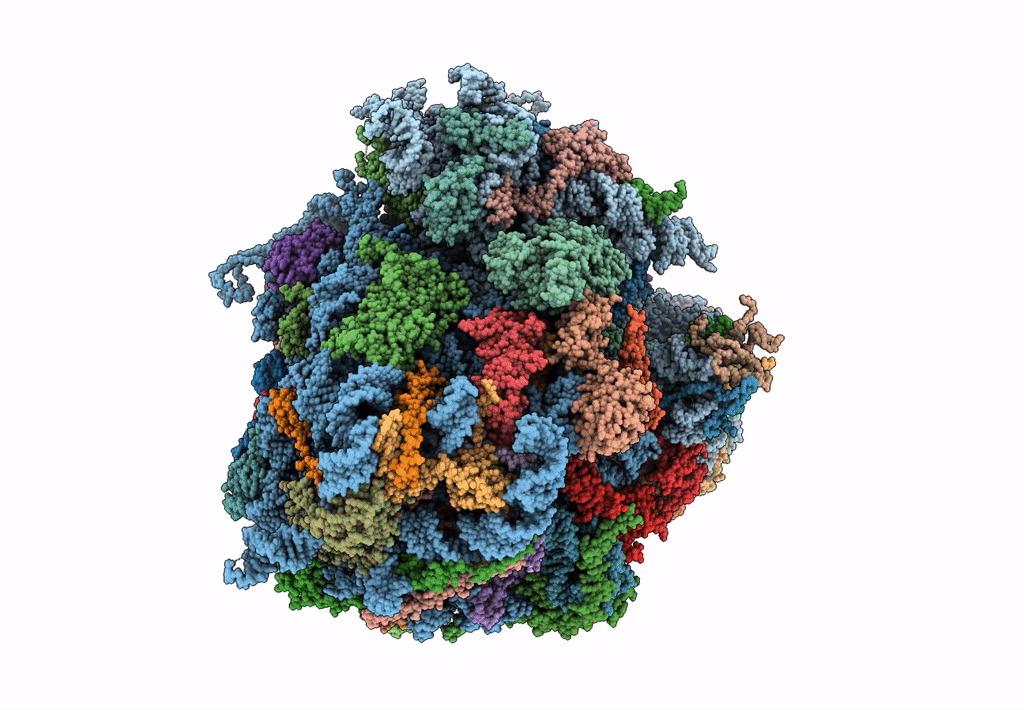
Saccharomyces cerevisiae 80S ribosome bound with elongation factor eEF2-GDP-sordarin and Taura Syndrome Virus IRES, Structure II (mid-rotated 40S subunit)
Biological Source:
Source Organism(s):
Taura syndrome virus (Taxon ID: 142102)
Saccharomyces cerevisiae (Taxon ID: 4932)
Saccharomyces cerevisiae (Taxon ID: 4932)
Method Details:
Experimental Method:
Resolution:
3.50 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE


