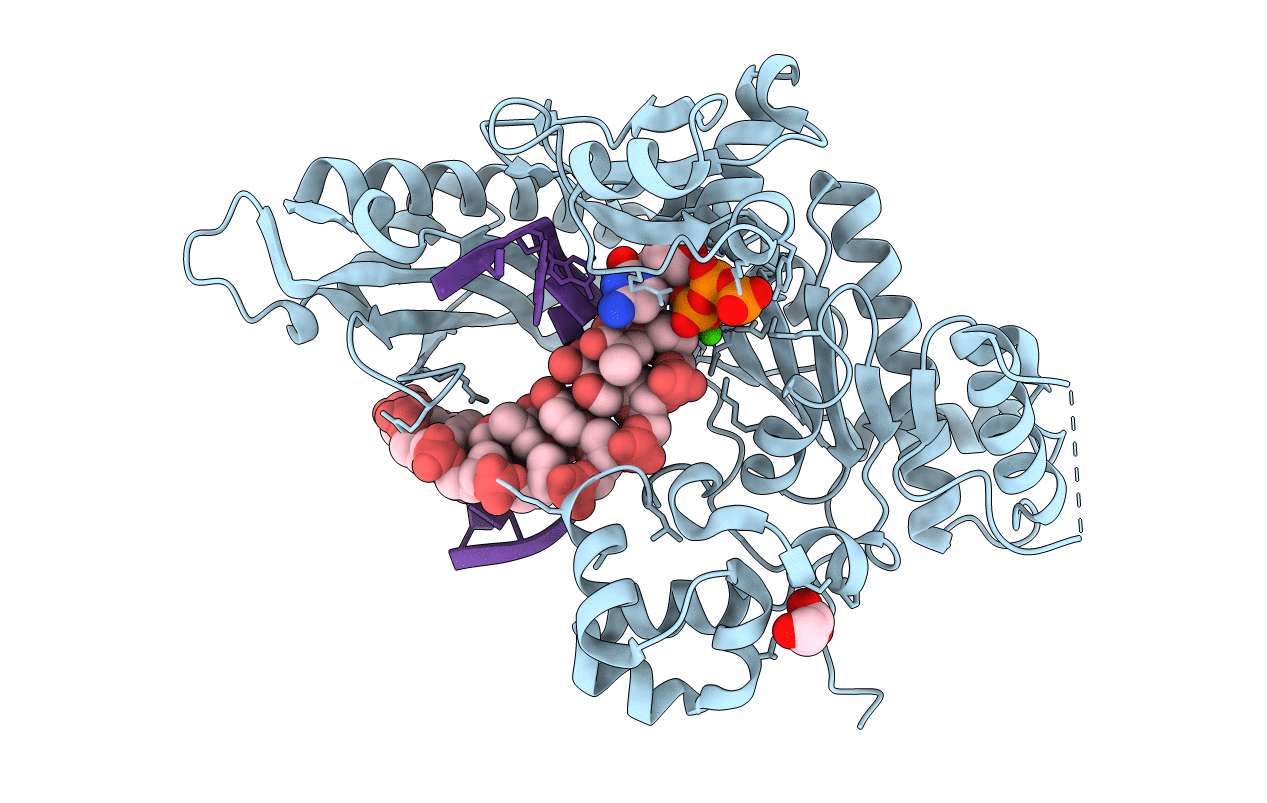
Deposition Date
2016-05-10
Release Date
2016-09-07
Last Version Date
2023-09-27
Entry Detail
PDB ID:
5JUM
Keywords:
Title:
Crystal Structure of Human DNA Polymerase Eta Inserting dCTP Opposite N-(2'-deoxyguanosin-8- yl)-3-aminobenzanthrone (C8-dG-ABA)
Biological Source:
Source Organism:
Homo sapiens (Taxon ID: 9606)
synthetic construct (Taxon ID: 32630)
synthetic construct (Taxon ID: 32630)
Host Organism:
Method Details:
Experimental Method:
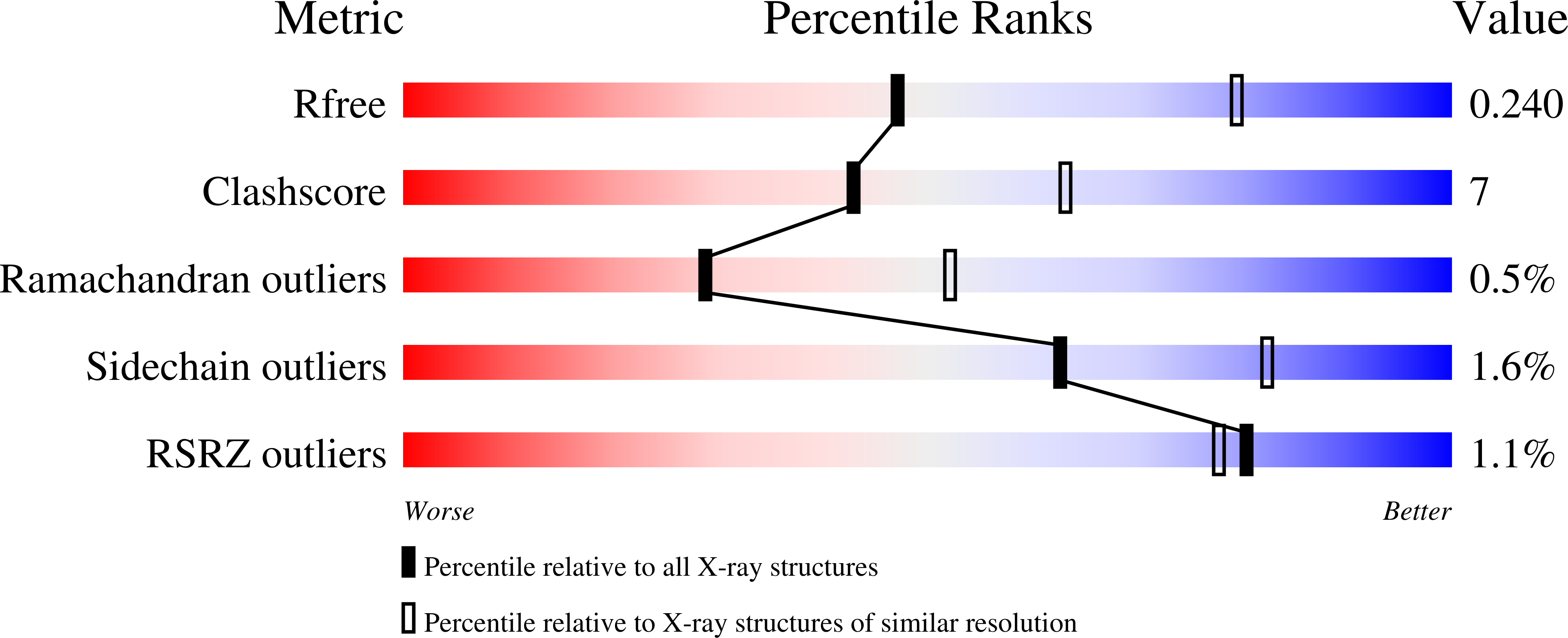
Resolution:
2.60 Å
R-Value Free:
0.23
R-Value Work:
0.16
R-Value Observed:
0.16
Space Group:
P 61