Deposition Date
2016-05-07
Release Date
2016-07-13
Last Version Date
2024-10-16
Entry Detail
PDB ID:
5JS9
Keywords:
Title:
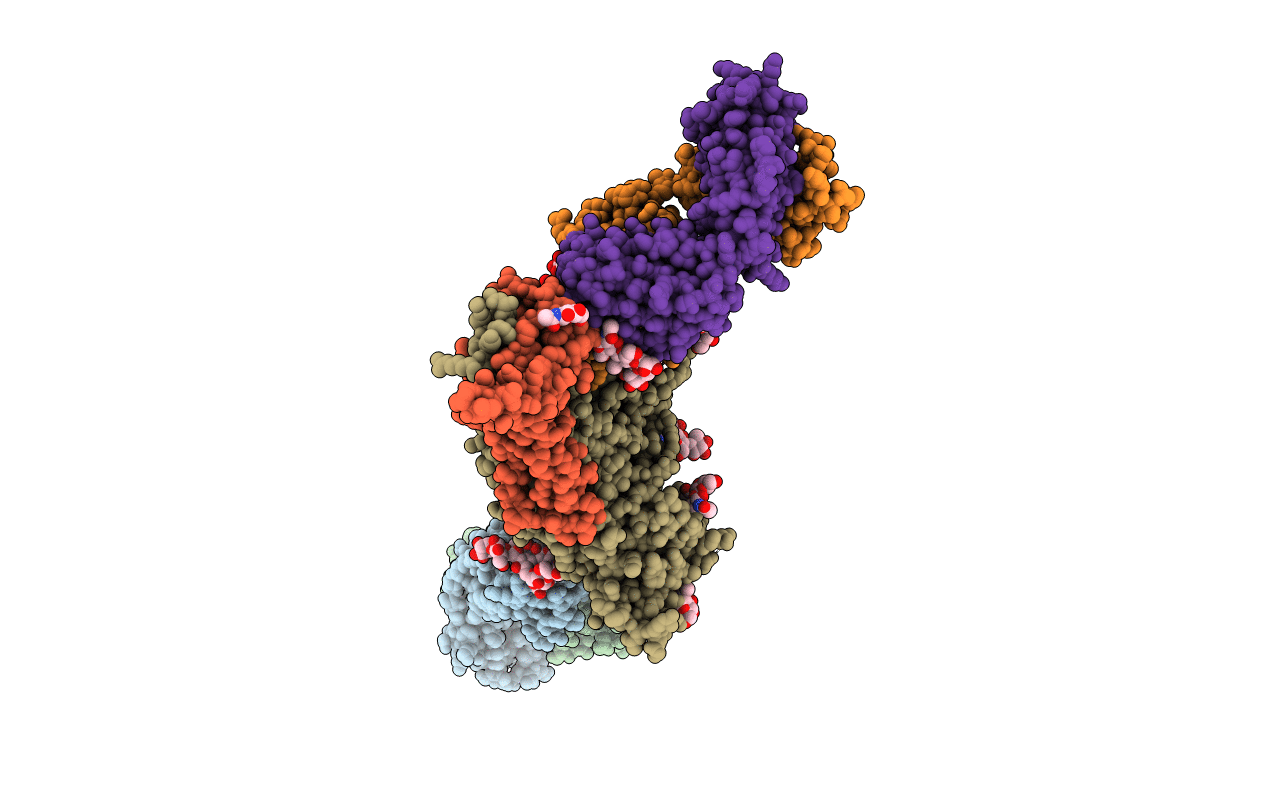
Uncleaved prefusion optimized gp140 trimer with an engineered 8-residue HR1 turn bound to broadly neutralizing antibodies 8ANC195 and PGT128
Biological Source:
Source Organism(s):
Homo sapiens (Taxon ID: 9606)
Human immunodeficiency virus 1 (Taxon ID: 11676)
Human immunodeficiency virus 1 (Taxon ID: 11676)
Expression System(s):
Method Details:
Experimental Method:
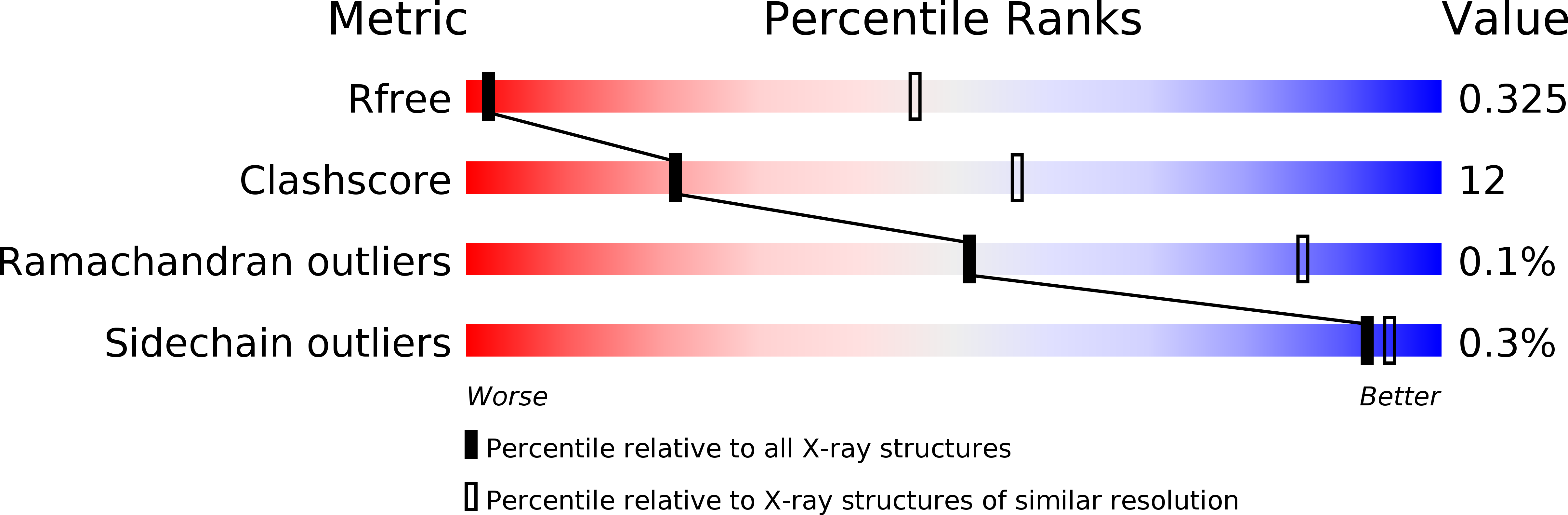
Resolution:
6.92 Å
R-Value Free:
0.32
R-Value Work:
0.28
R-Value Observed:
0.28
Space Group:
I 2 3