Deposition Date
2016-04-18
Release Date
2016-07-13
Last Version Date
2024-01-10
Entry Detail
PDB ID:
5JEA
Keywords:
Title:
Structure of a cytoplasmic 11-subunit RNA exosome complex including Ski7, bound to RNA
Biological Source:
Source Organism:
Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Taxon ID: 559292)
Enterobacteria phage T4 (Taxon ID: 10665)
synthetic construct (Taxon ID: 32630)
Enterobacteria phage T4 (Taxon ID: 10665)
synthetic construct (Taxon ID: 32630)
Host Organism:
Method Details:
Experimental Method:
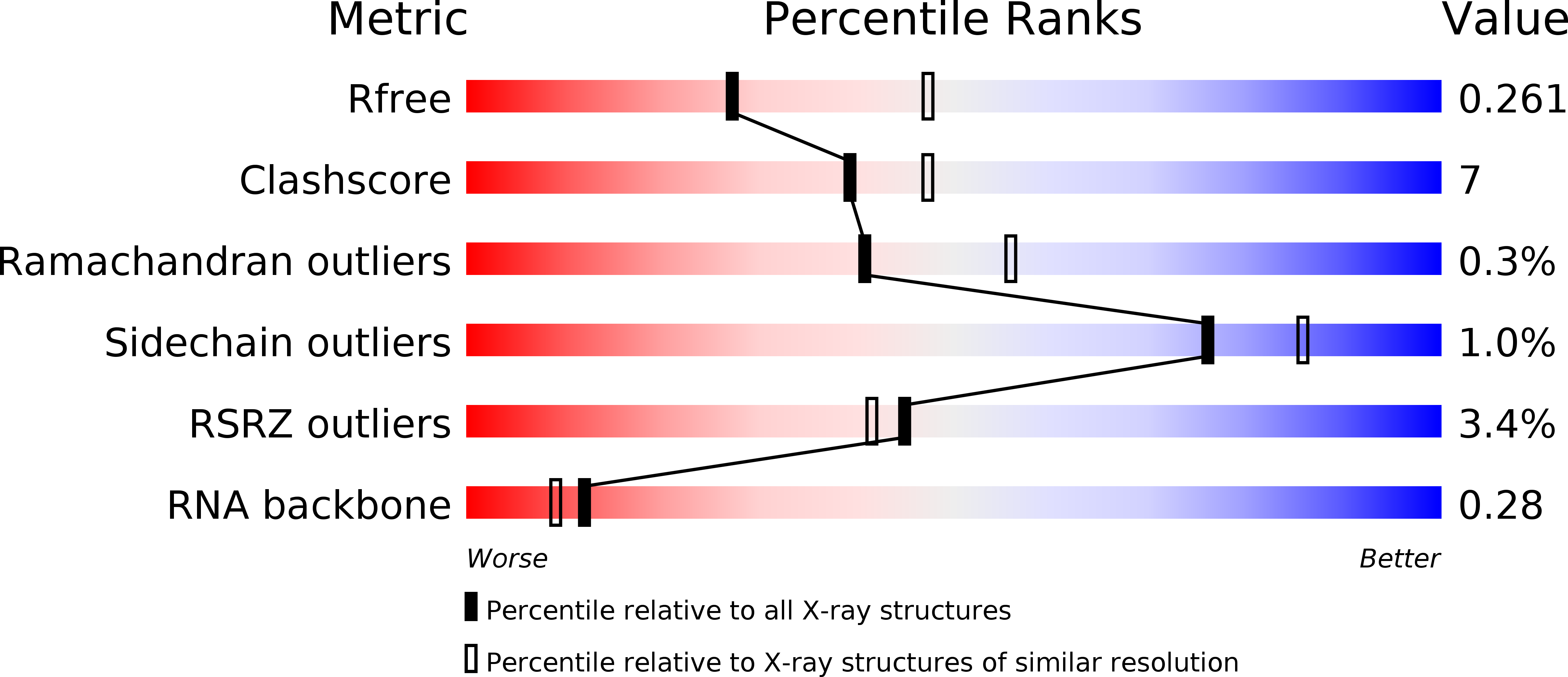
Resolution:
2.65 Å
R-Value Free:
0.25
R-Value Work:
0.21
R-Value Observed:
0.21
Space Group:
P 21 21 21