Deposition Date
2016-03-29
Release Date
2016-06-15
Last Version Date
2023-09-27
Entry Detail
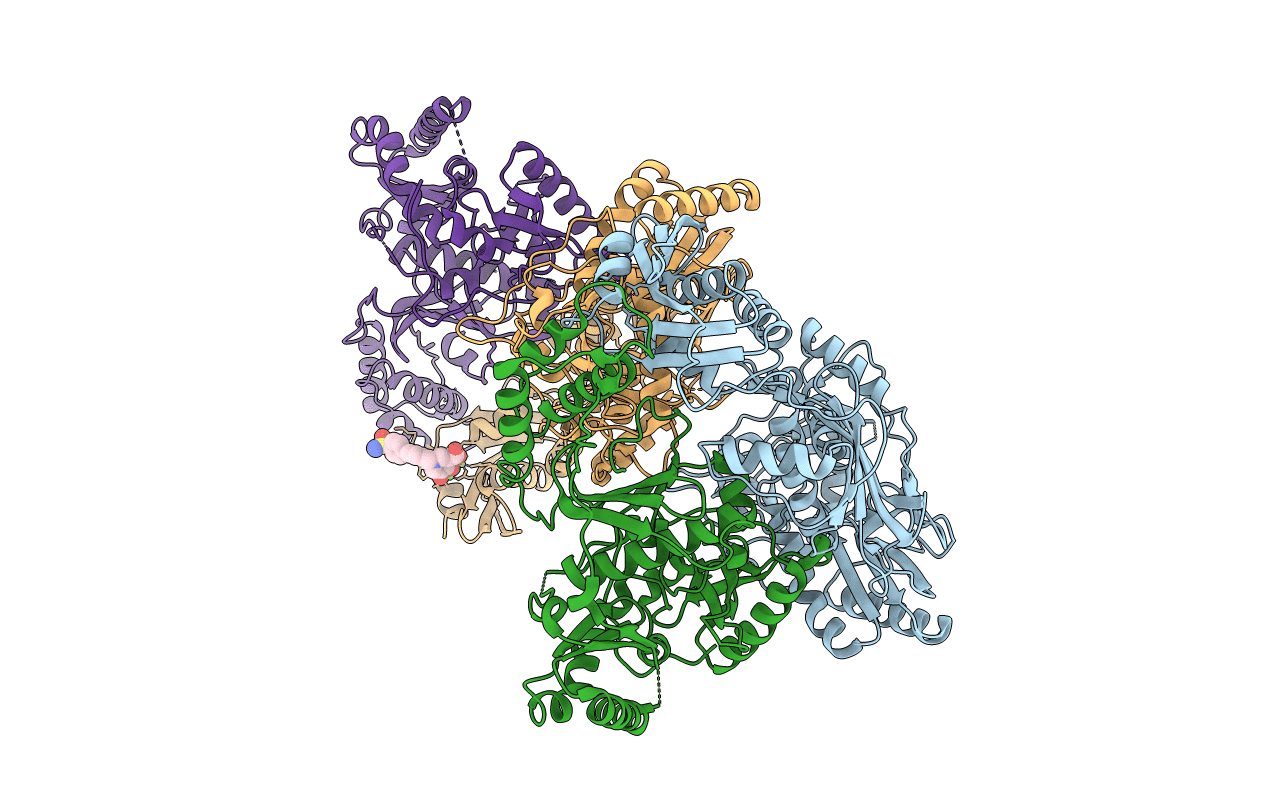
PDB ID:
5J1E
Keywords:
Title:
Crystal Structure of a Hydroxypyridone Carboxylic Acid Active-Site RNase H Inhibitor in Complex with HIV Reverse Transcriptase
Biological Source:
Source Organism:
Host Organism:
Method Details:
Experimental Method:
Resolution:
2.90 Å
R-Value Free:
0.29
R-Value Work:
0.25
R-Value Observed:
0.25
Space Group:
P 1


