Deposition Date
2016-03-26
Release Date
2016-12-14
Last Version Date
2024-01-10
Entry Detail
PDB ID:
5J01
Keywords:
Title:
Structure of the lariat form of a chimeric derivative of the Oceanobacillus iheyensis group II intron in the presence of NH4+ and MG2+.
Biological Source:
Source Organism(s):
Oceanobacillus iheyensis (Taxon ID: 182710)
Method Details:
Experimental Method:
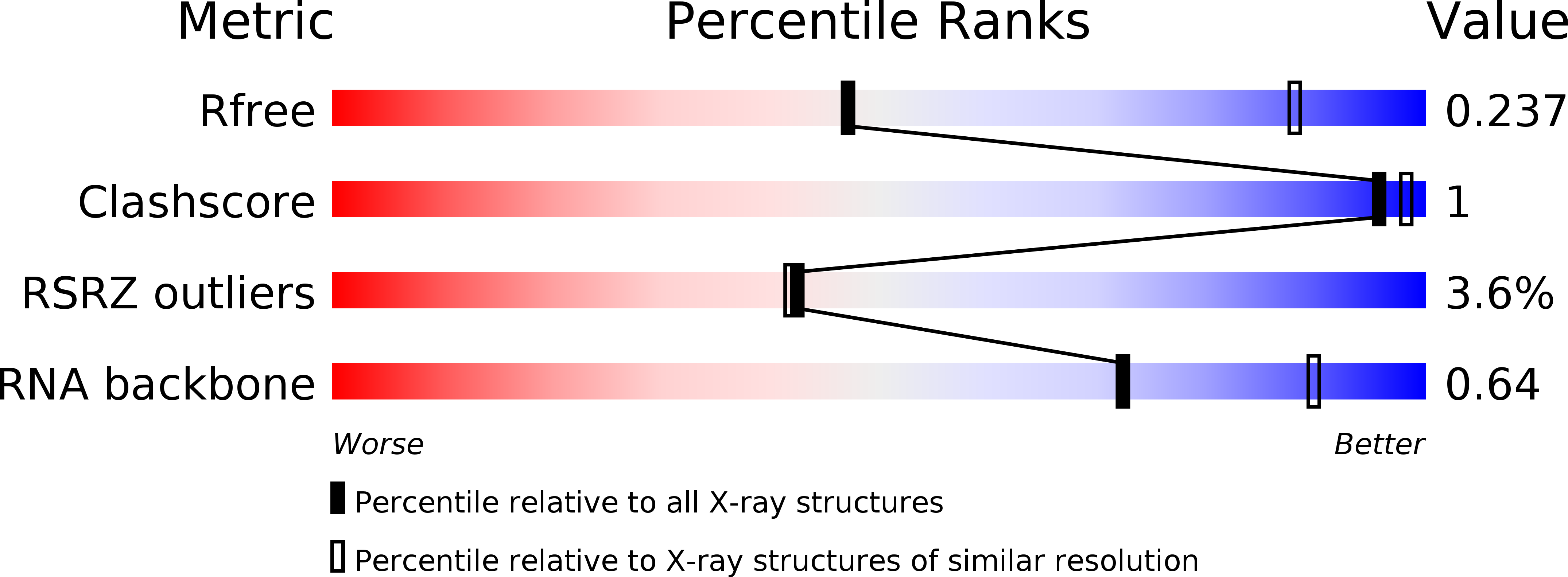
Resolution:
3.39 Å
R-Value Free:
0.23
R-Value Work:
0.20
R-Value Observed:
0.20
Space Group:
P 21 21 2