Deposition Date
2016-03-24
Release Date
2017-01-11
Last Version Date
2023-11-08
Entry Detail
PDB ID:
5IY5
Keywords:
Title:
Electron transfer complex of cytochrome c and cytochrome c oxidase at 2.0 angstrom resolution
Biological Source:
Source Organism:
Bos taurus (Taxon ID: 9913)
Equus caballus (Taxon ID: 9796)
Equus caballus (Taxon ID: 9796)
Method Details:
Experimental Method:
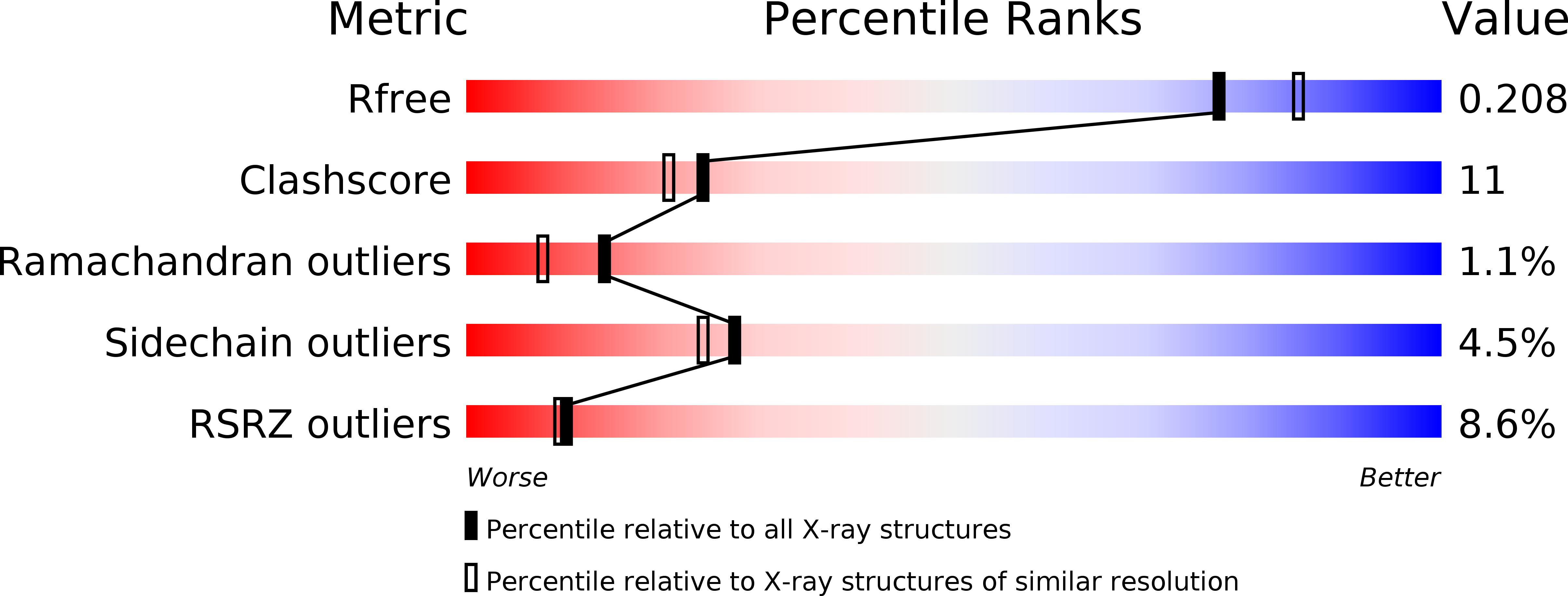
Resolution:
2.00 Å
R-Value Free:
0.20
R-Value Work:
0.16
R-Value Observed:
0.16
Space Group:
P 1 21 1