Deposition Date
2016-02-28
Release Date
2016-06-29
Last Version Date
2024-10-30
Entry Detail
PDB ID:
5IH2
Keywords:
Title:
Structure, thermodynamics, and the role of conformational dynamics in the interactions between the N-terminal SH3 domain of CrkII and proline-rich motifs in cAbl
Biological Source:
Source Organism(s):
Mus musculus (Taxon ID: 10090)
Endothia gyrosa (Taxon ID: 40263)
Endothia gyrosa (Taxon ID: 40263)
Expression System(s):
Method Details:
Experimental Method:
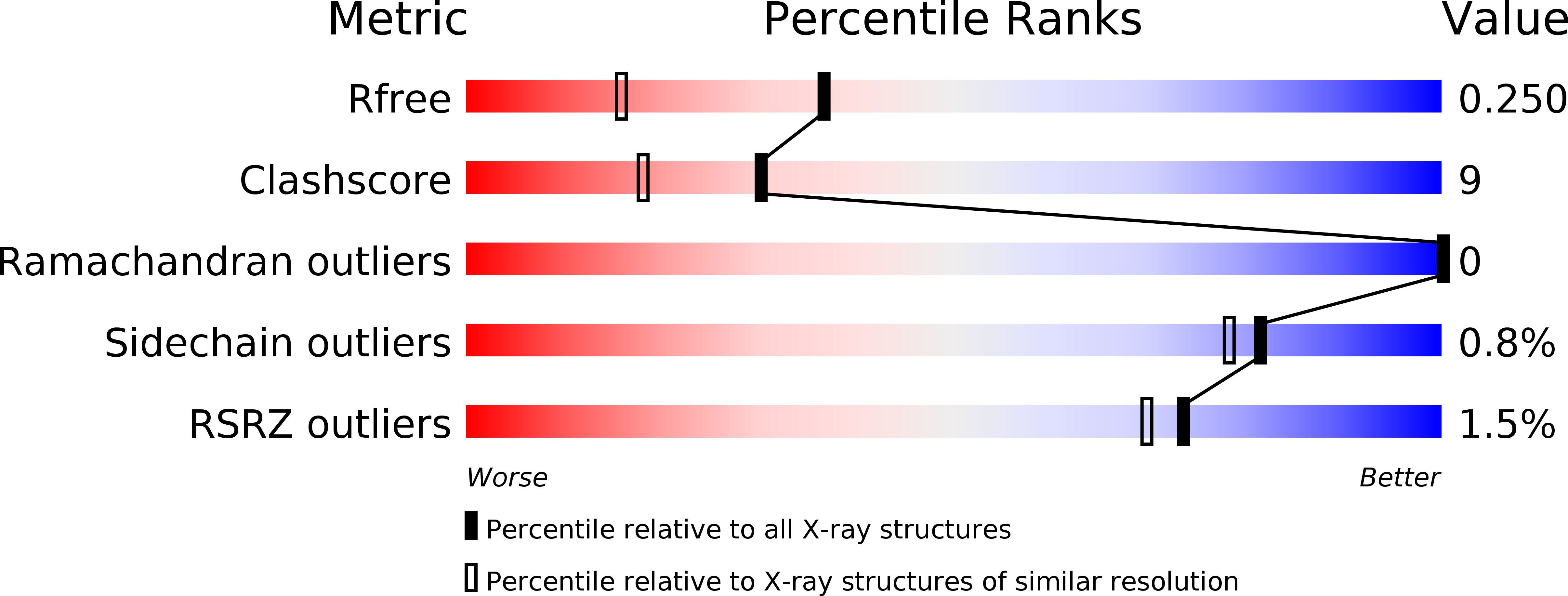
Resolution:
1.80 Å
R-Value Free:
0.24
R-Value Work:
0.16
R-Value Observed:
0.17
Space Group:
P 1 21 1