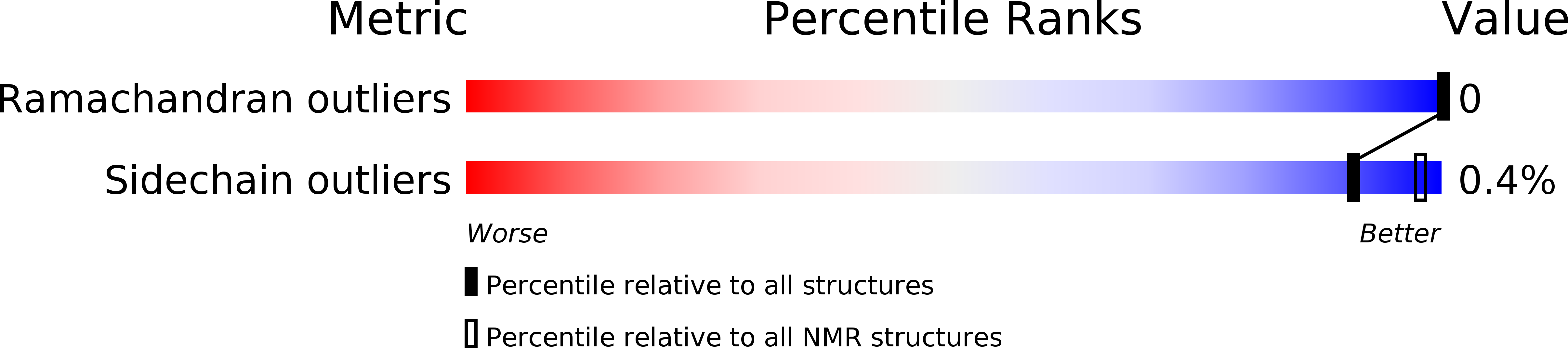
Deposition Date
2016-02-05
Release Date
2016-03-30
Last Version Date
2024-05-15
Entry Detail
PDB ID:
5I1R
Keywords:
Title:
Quantitative characterization of configurational space sampled by HIV-1 nucleocapsid using solution NMR and X-ray scattering
Biological Source:
Source Organism(s):
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
100
Conformers Submitted:
21
Selection Criteria:
structures with the lowest energy