Deposition Date
2016-02-01
Release Date
2017-02-01
Last Version Date
2023-11-08
Entry Detail
PDB ID:
5HYC
Keywords:
Title:
Structure based function annotation of a hypothetical protein MGG_01005 related to the development of rice blast fungus
Biological Source:
Source Organism(s):
Expression System(s):
Method Details:
Experimental Method:
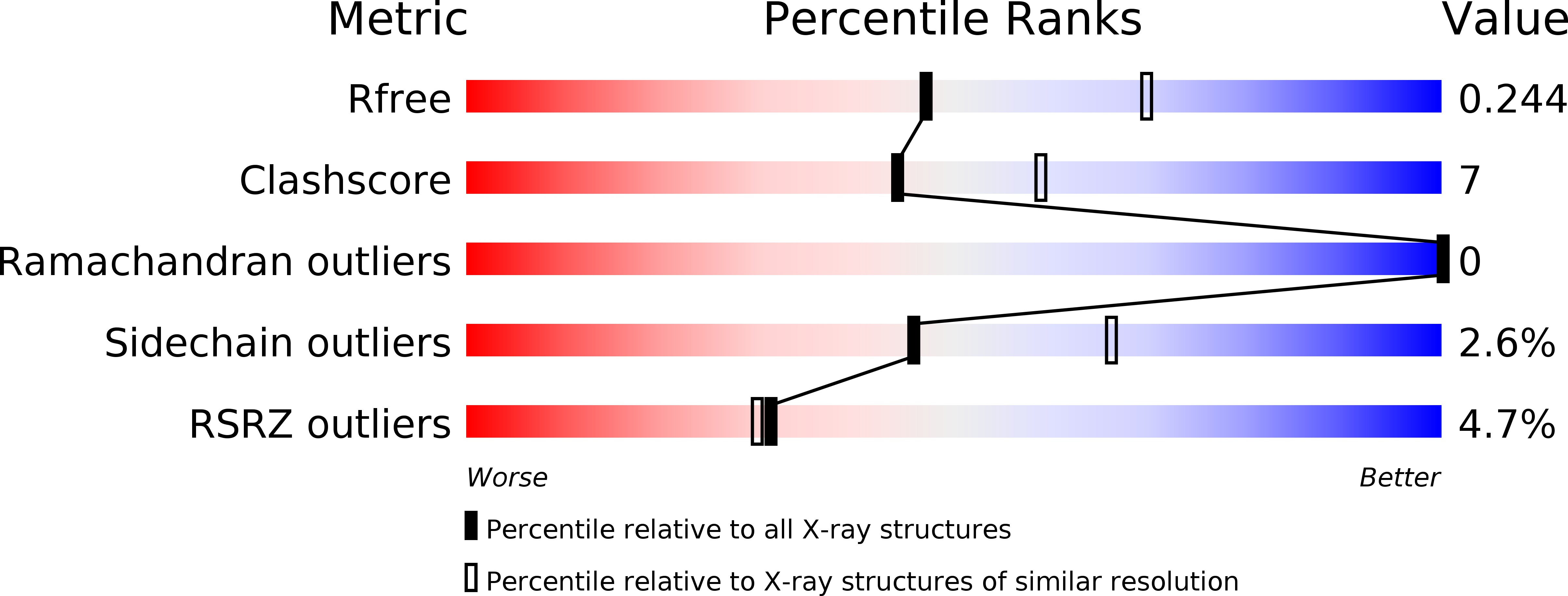
Resolution:
2.40 Å
R-Value Free:
0.23
R-Value Work:
0.20
R-Value Observed:
0.20
Space Group:
I 41 2 2