Deposition Date
2015-12-28
Release Date
2016-02-17
Last Version Date
2023-09-27
Entry Detail
PDB ID:
5H9F
Keywords:
Title:
Crystal structure of E. coli Cascade bound to a PAM-containing dsDNA target at 2.45 angstrom resolution.
Biological Source:
Source Organism(s):
Escherichia coli (strain K12) (Taxon ID: 83333)
Escherichia coli (Taxon ID: 562)
Escherichia coli (Taxon ID: 562)
Expression System(s):
Method Details:
Experimental Method:
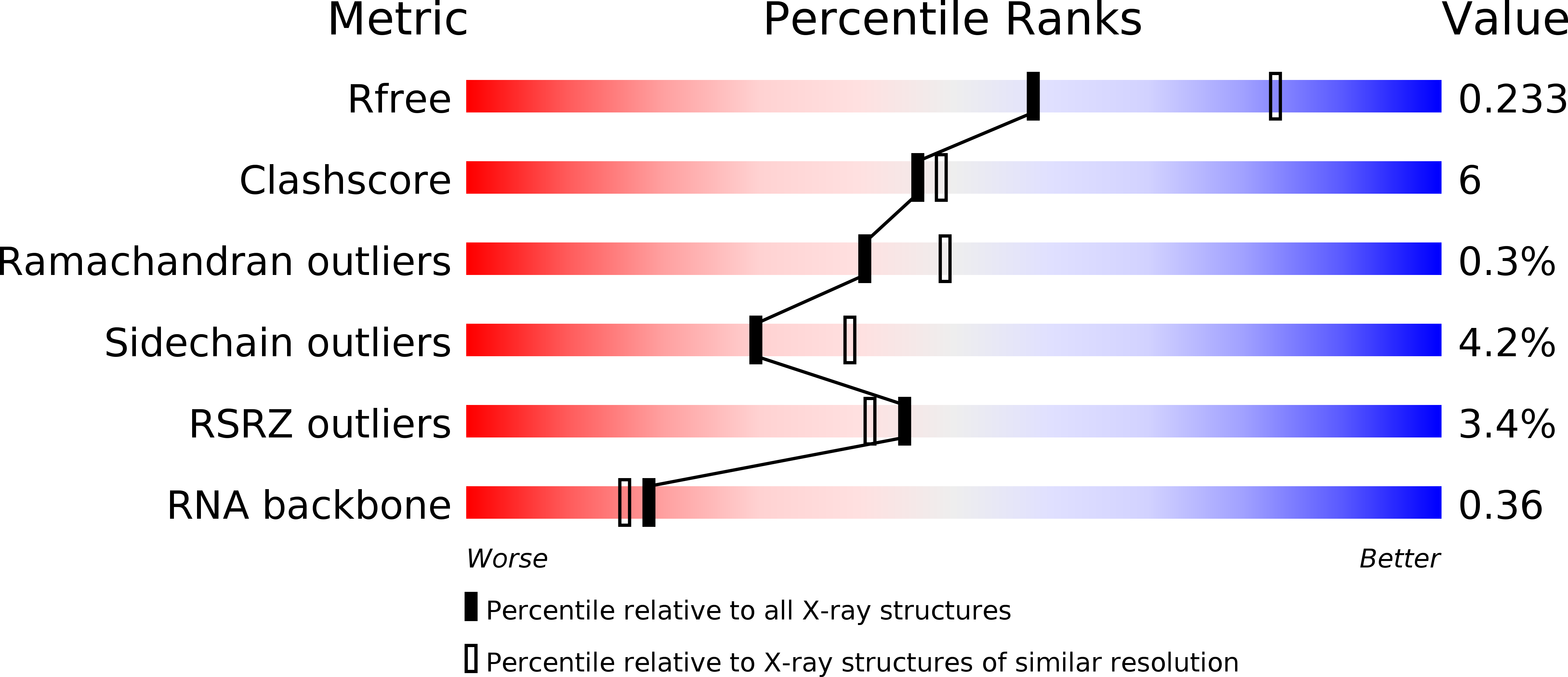
Resolution:
2.45 Å
R-Value Free:
0.23
R-Value Work:
0.20
R-Value Observed:
0.20
Space Group:
P 2 21 21