Deposition Date
2015-11-26
Release Date
2015-12-16
Last Version Date
2024-01-10
Entry Detail
PDB ID:
5EZK
Keywords:
Title:
RNA polymerase model placed by Molecular replacement into X-ray diffraction map of DNA-bound RNA Polymerase-Sigma 54 holoenzyme complex.
Biological Source:
Source Organism(s):
Escherichia coli (Taxon ID: 562)
Expression System(s):
Method Details:
Experimental Method:
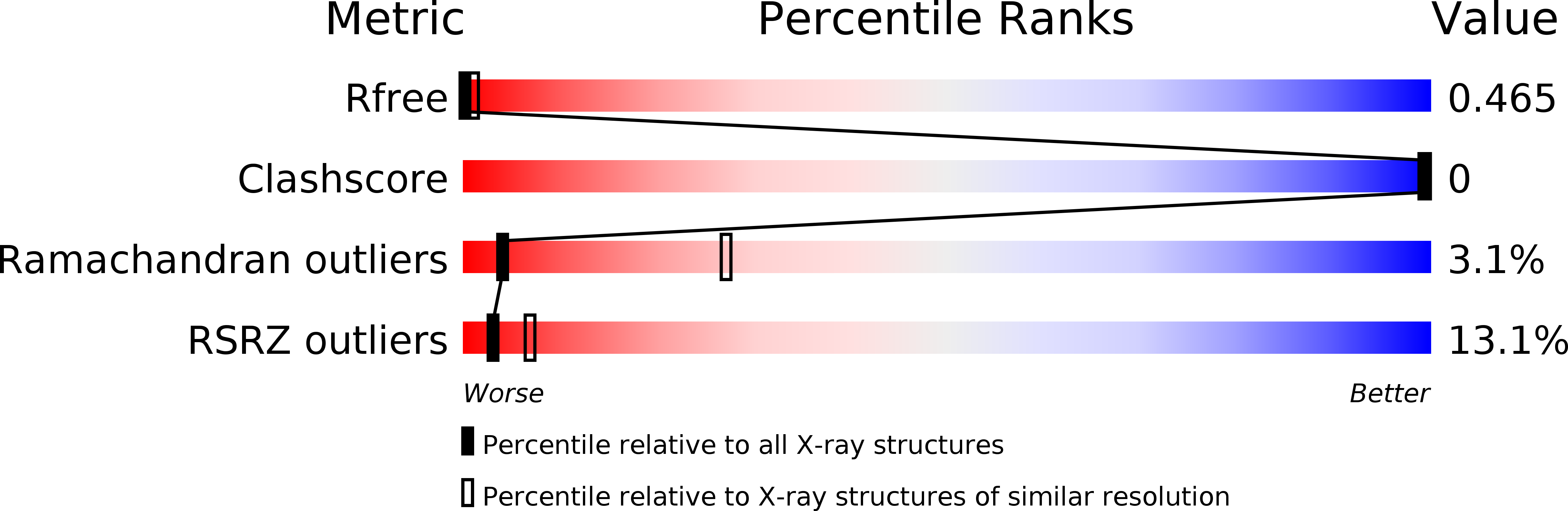
Resolution:
8.50 Å
R-Value Free:
0.47
R-Value Work:
0.47
R-Value Observed:
0.47
Space Group:
P 63